miRNA library prep
miRNA library prep for Mcap developmental time series Hawaii 2023
This post details the info about the miRNA library prep steps for the M. capitata developmental time series experiment in Hawaii 2023. The github for that project is linked here. I’m using the Zymo-Seq miRNA library kit for small RNA library prep (I got the 12 prep kit). See Zymo’s protocol here.
I’ll be using samples M9, M13, M23, M35, M52 and M85. M9 was extracted on 2/22/24, and all of the other samples were extracted on 2/10/24. These samples, along with M60 and M72 (prepped on 3/4/24), will be sent for sequencing. The samples represent n=1 from each time point during M. capitata development. I will use 150-200 ng of RNA from each sample.
Here’s the miRNA library prep workflow:
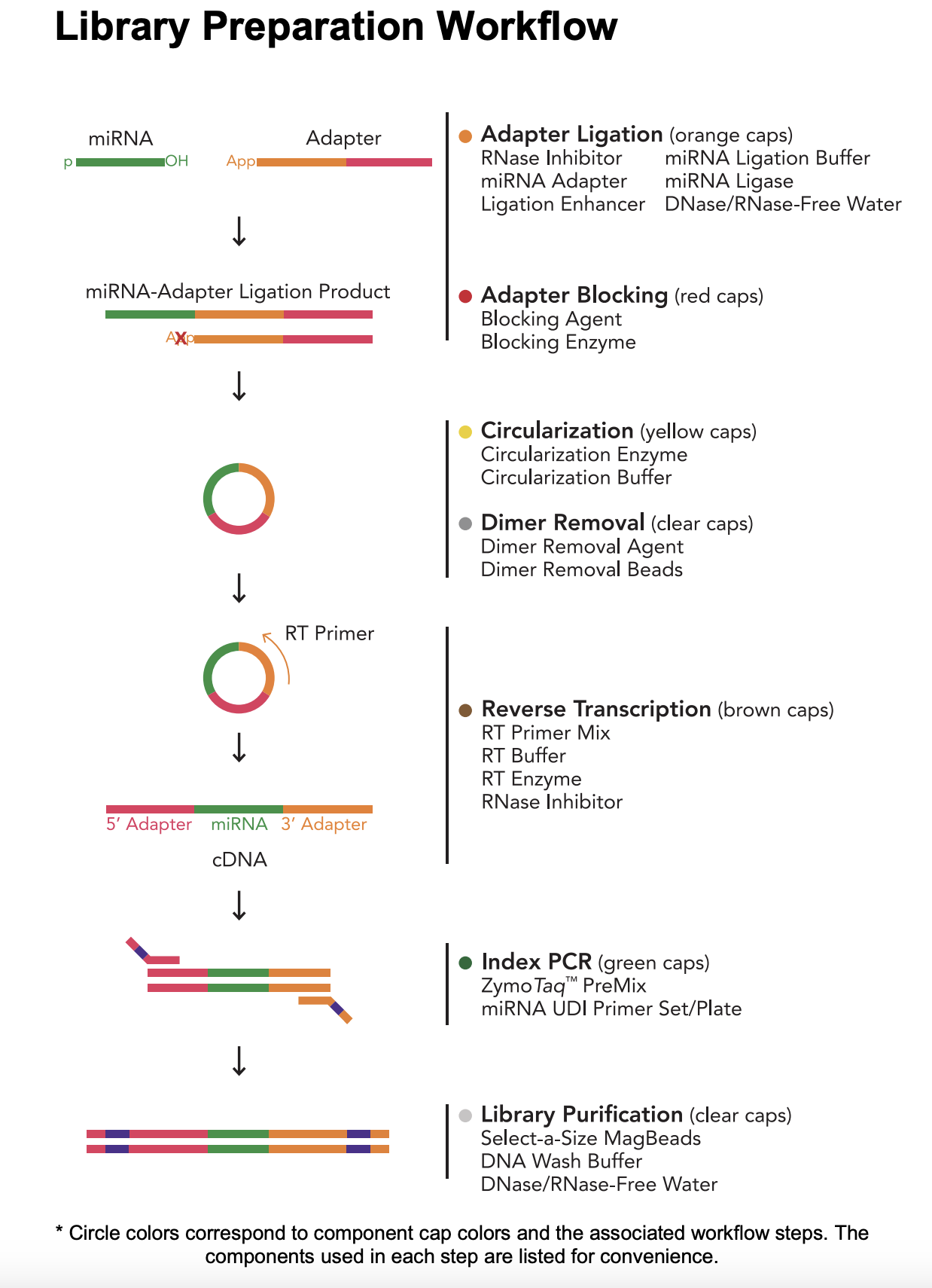
Materials
- Zymo-Seq miRNA library kit
- PCR tubes
- Thermocycler
- Heating block
- Mini centrifuge
- Bench top centrifuge
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- Kit contents
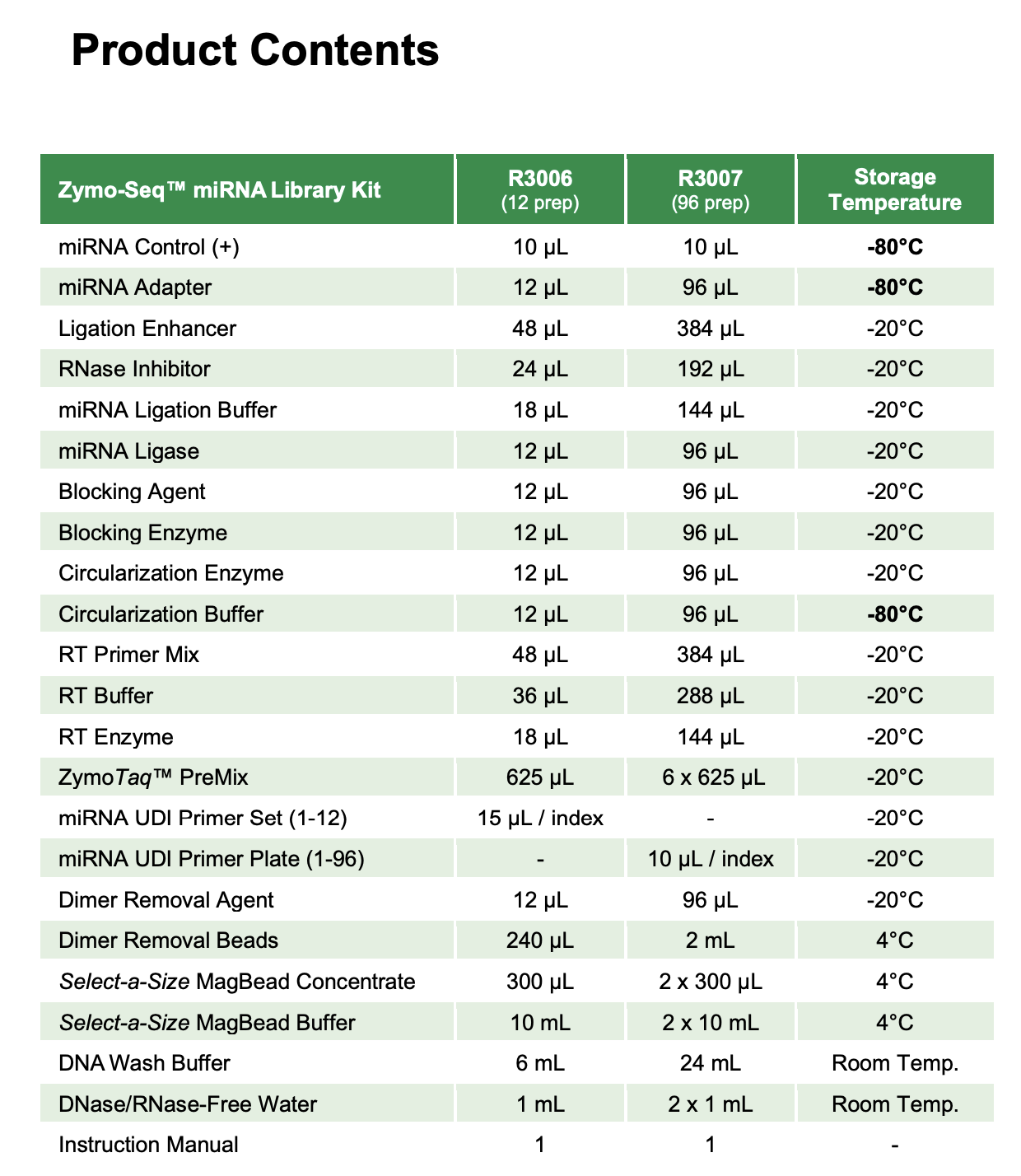
Materials
Protocol was followed according to this post. A few differences (and errors):
- I changed the input RNA amount to ~150-200 ng
- I increased the number of amplification cycles from 16 to 22
- After the 65°C hold in Section 1, the thermocycler was supposed to go into a 4°C hold. Instead, it went into a 65°C for 45 minutes.
- In Section 3, I added 4 uL of RT Buffer to samples instead of 4 uL of RT Primer. After I realized this error, I added 4 uL of RT Primer and ran Steps 1-2 on the thermocycler. I made the RT master mix without the RT Buffer (as I did not have any left). To make it, I combined 6.6 uL of RNase Inhibitor aand 9.9 uL RT enzyme, then added 2.75 uL to each sample.
- I did Sections 1-3 on 3/6/24, then stored overnight at -20°C. On 3/7/24, I did Section 4 and QC.
Both of the above errors likely ruined these libraries, which is very unfortunate because that was the last preps that I had for this kit.
QC
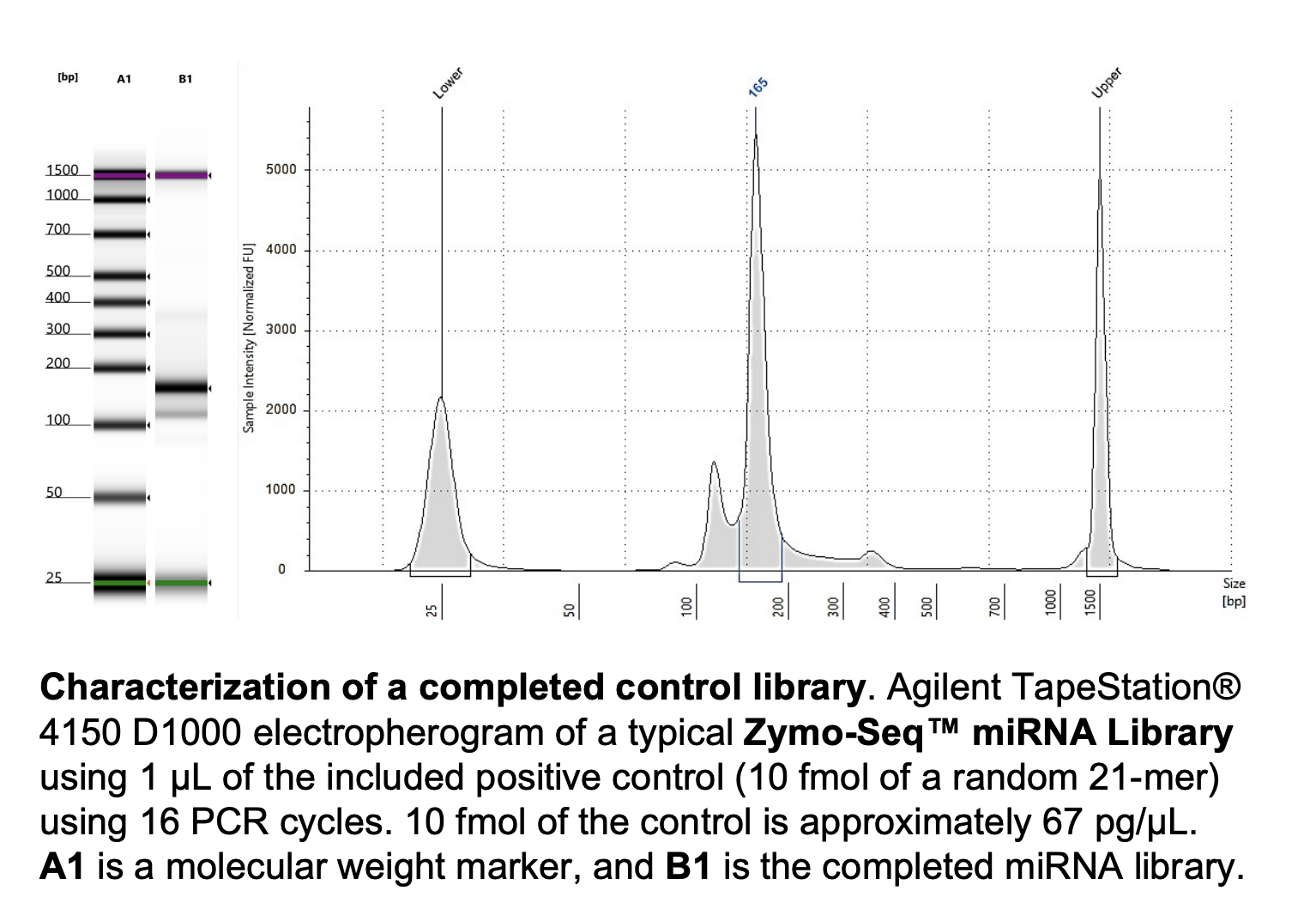
Run DNA Tapestation for visualize libraries. Here’s an example of what the library should look like on a Tapestation:
Here’s how my samples turned out on 3/7/24. See full report here.
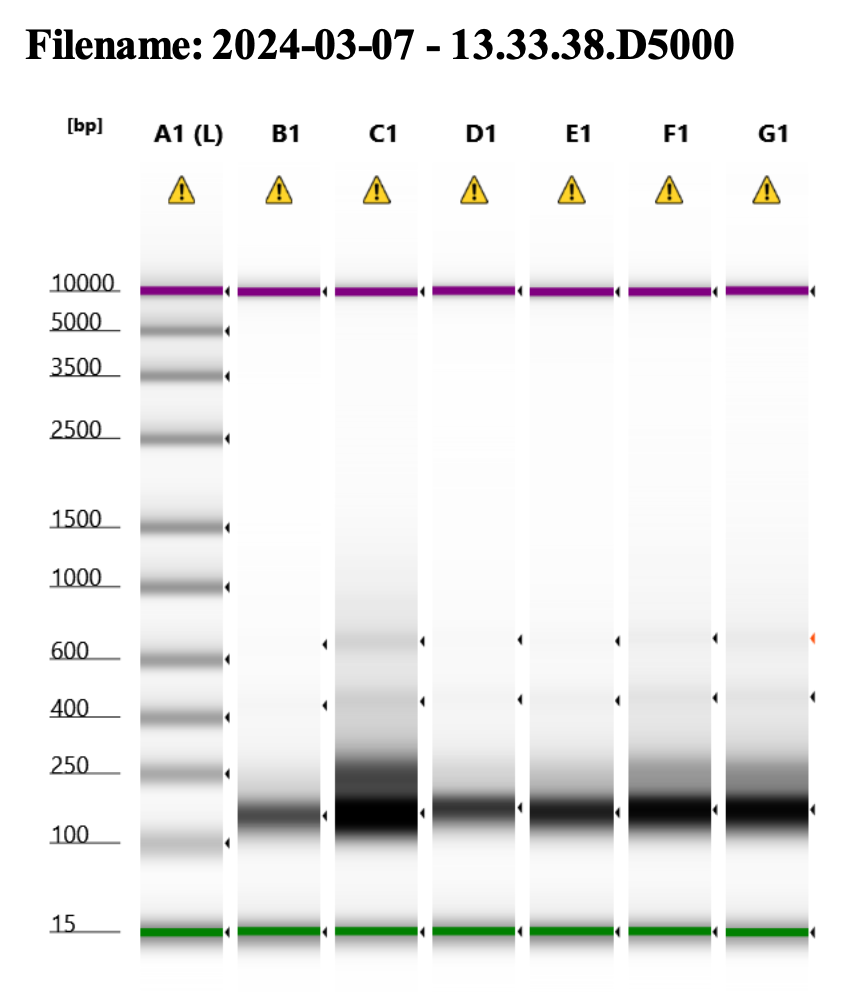
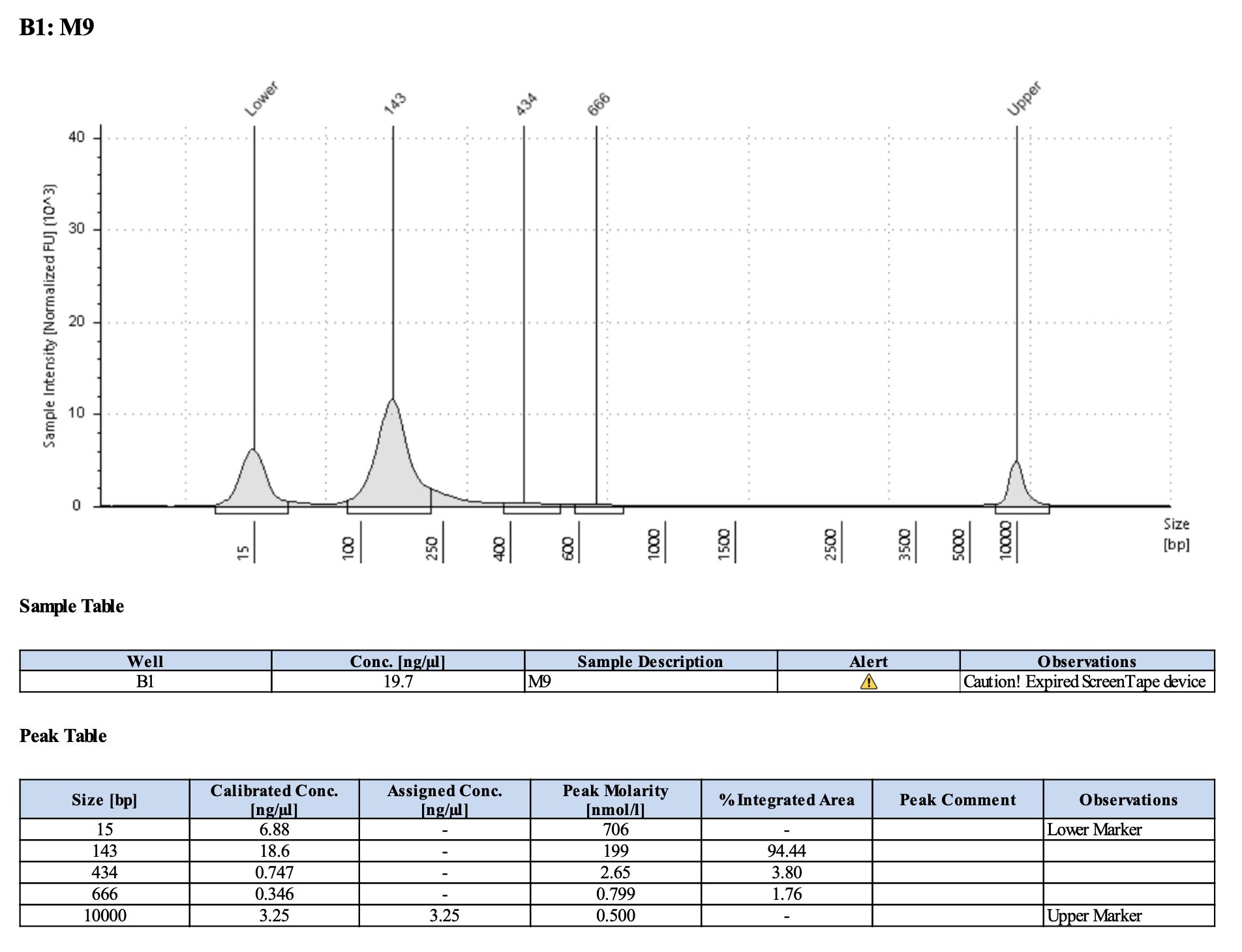
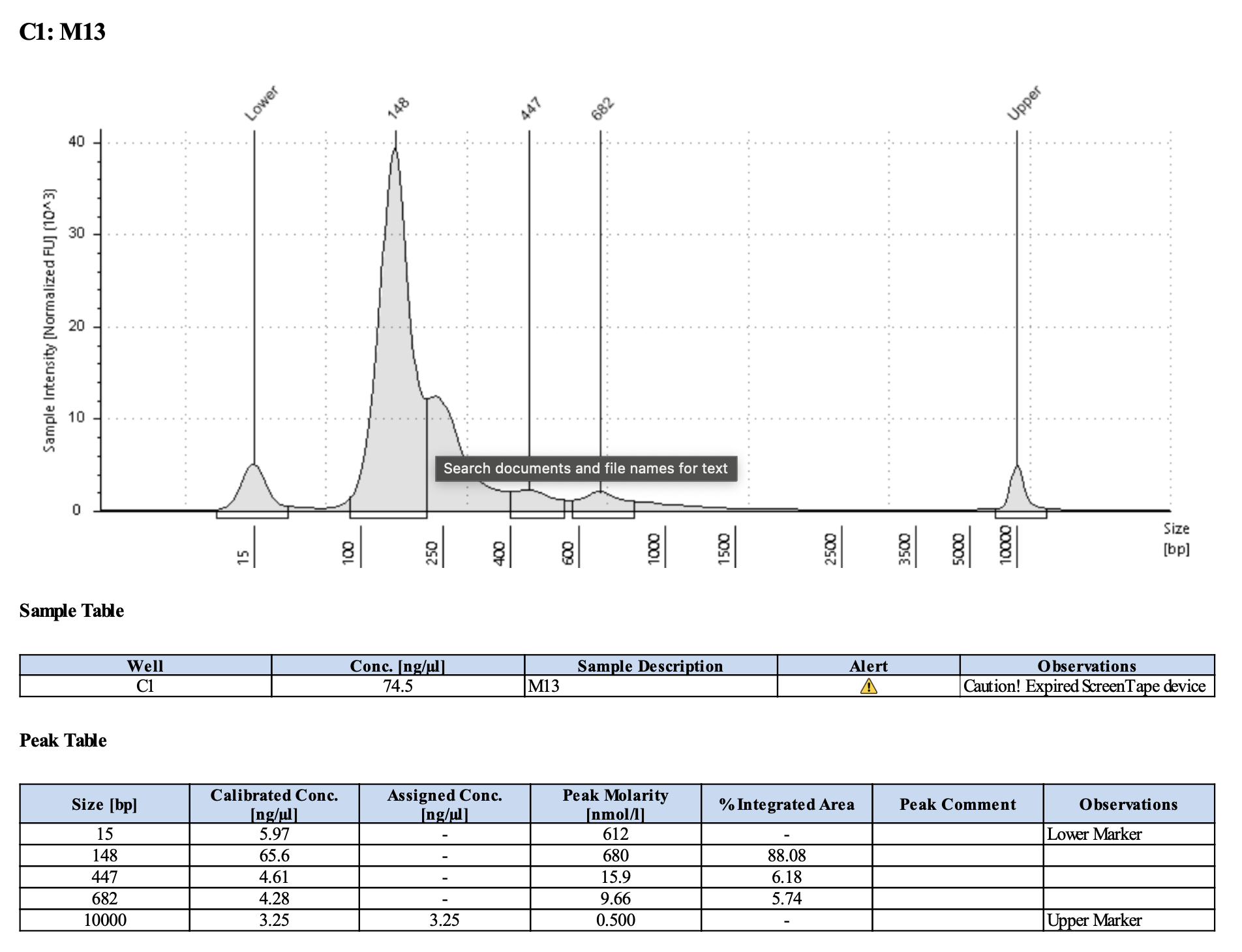
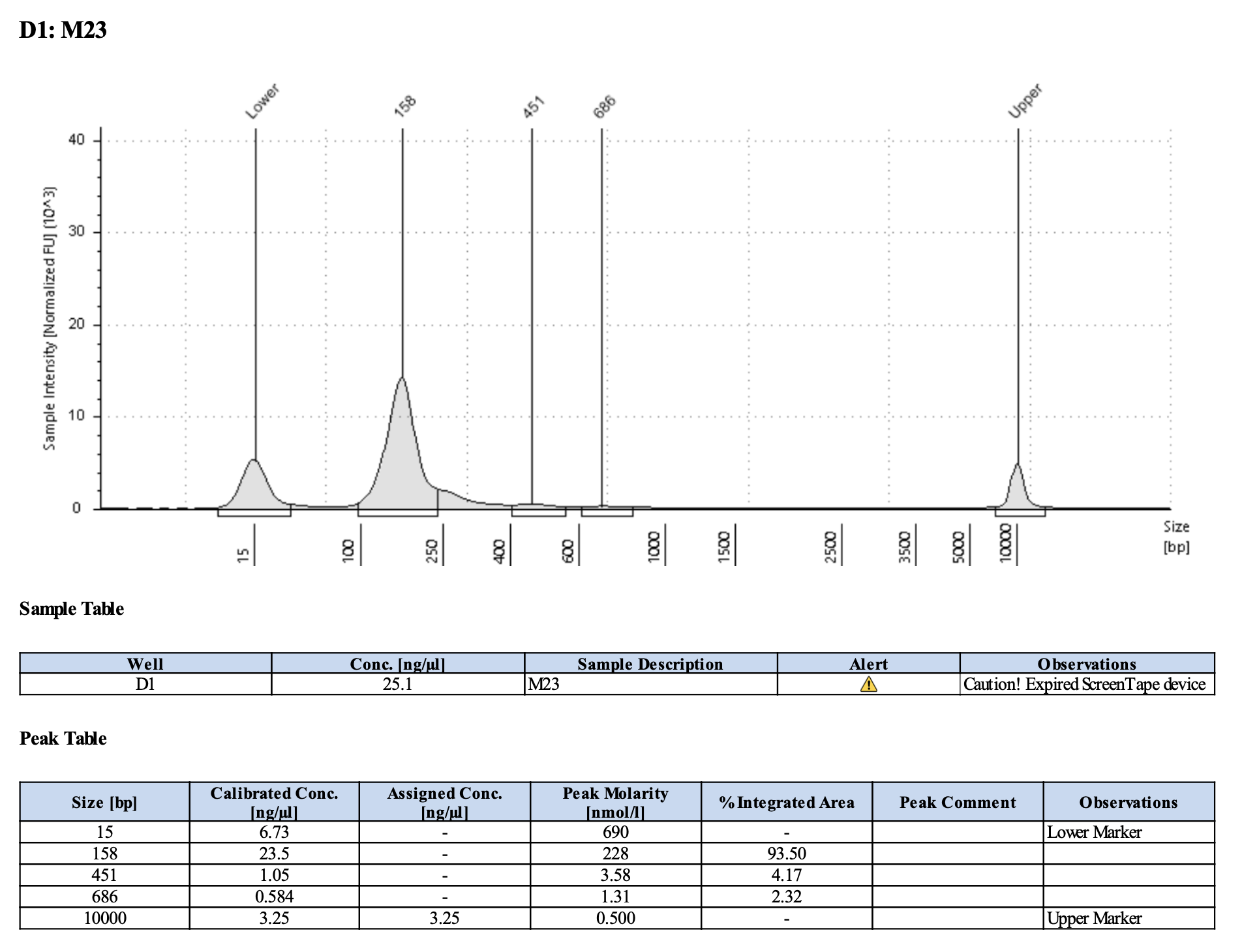
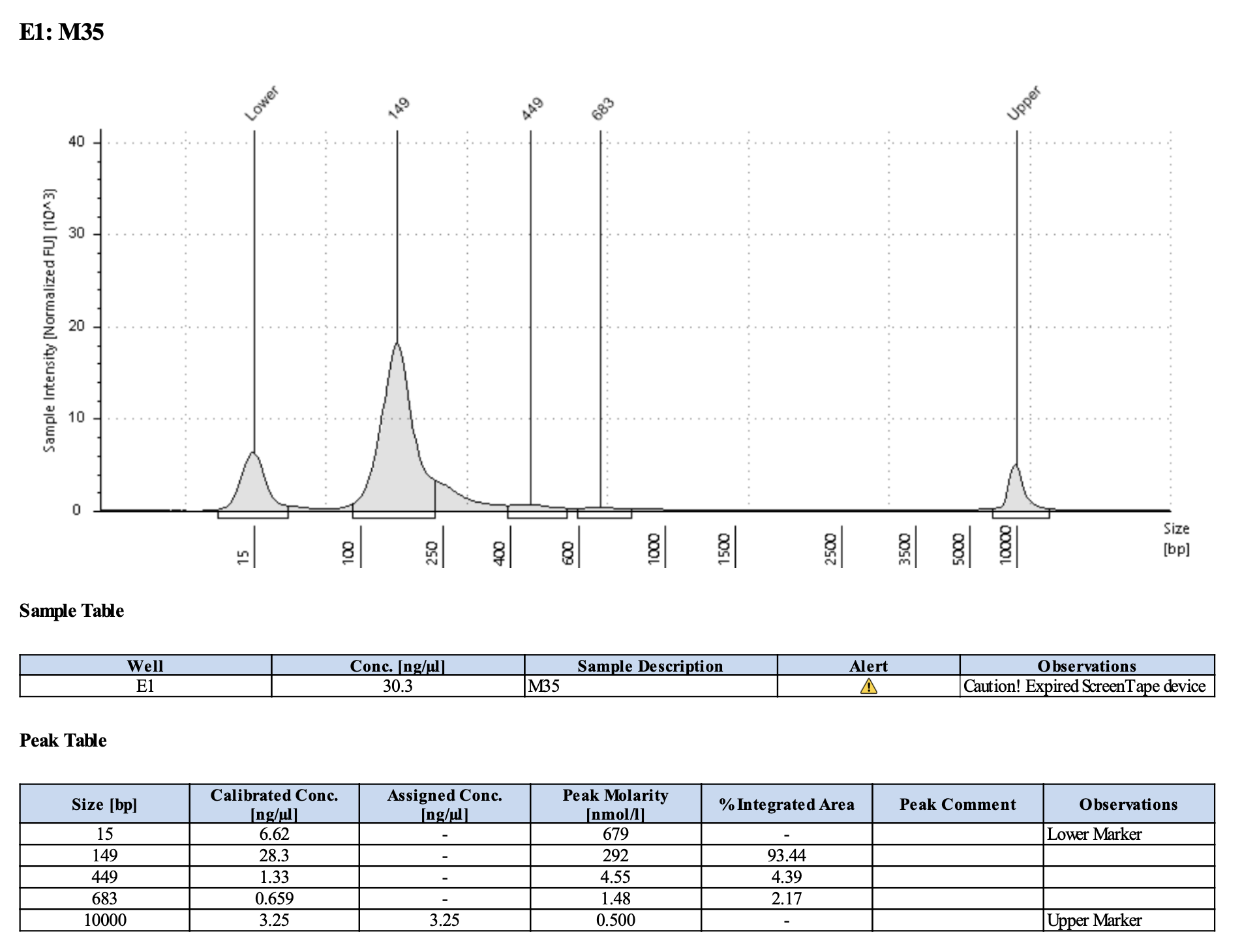
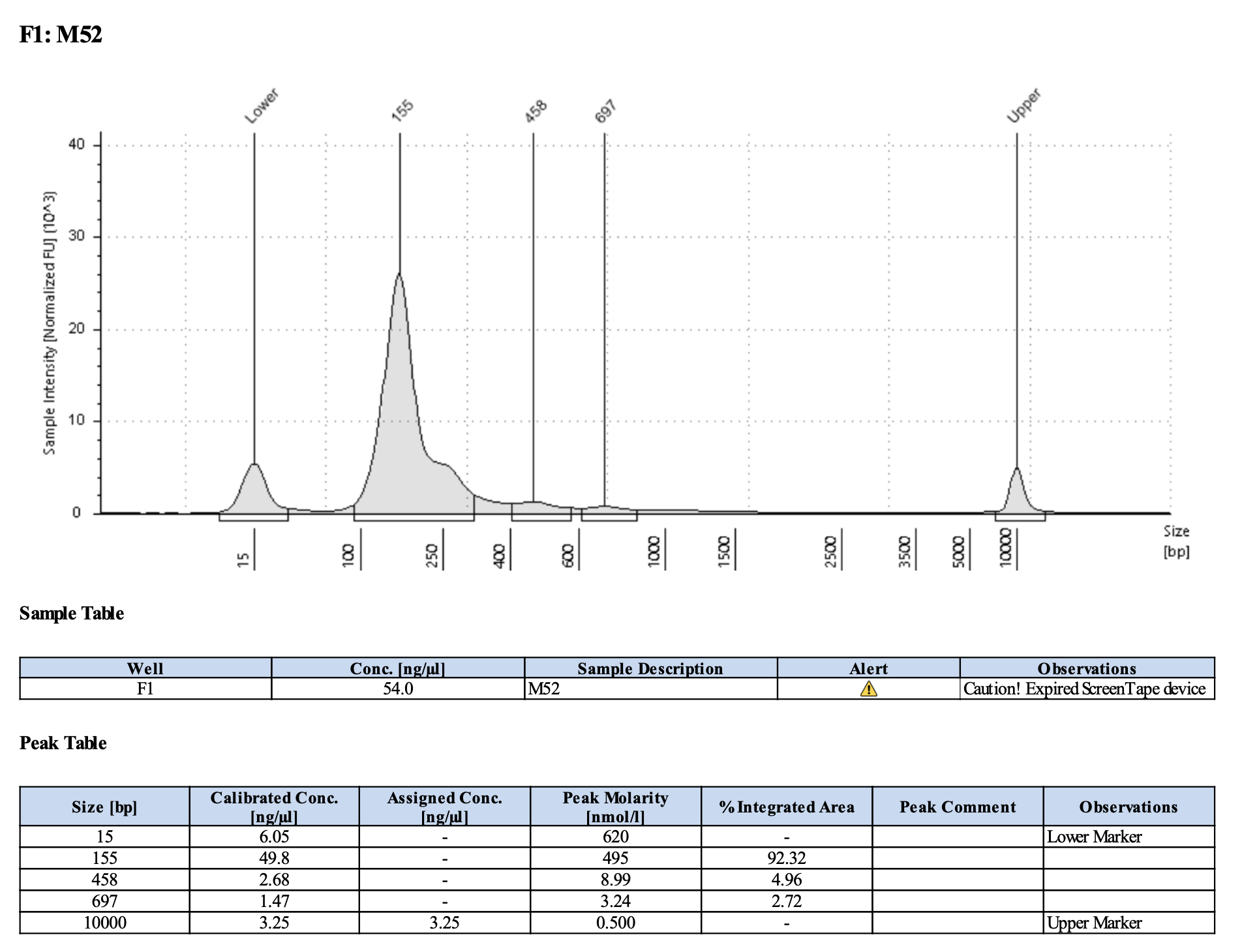
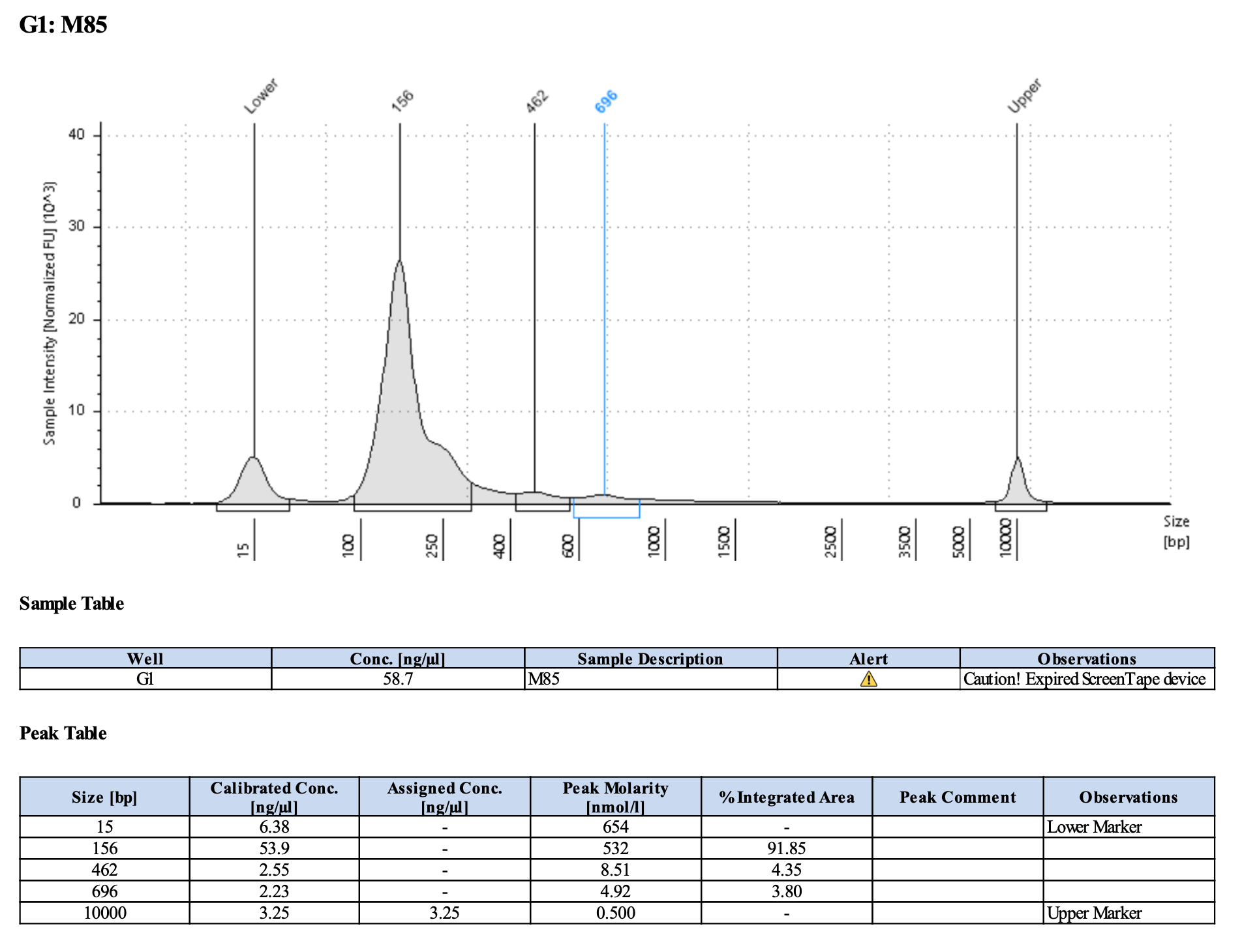
Wow I am shocked. I thought these libraries were ruined but they actually look very good!!!!! M9, M13, and M35 all have peaks around 145-148 bp, which isn’t ideal, as the Zymo protocol says that these might be dimers. But its hard to say because the bp that they recommend is 165 bp, which is very close to 148 bp. All look good to send for sequencing now!