miRNA library prep
miRNA library prep for Mcap developmental time series Hawaii 2023
This post details the info about the miRNA library prep steps for the M. capitata developmental time series experiment in Hawaii 2023. The github for that project is linked here. I’m using the Zymo-Seq miRNA library kit for small RNA library prep (I got the 12 prep kit). See Zymo’s protocol here.
After sequencing n=1 from each time point from the Mcap 2023 project, we decided to move forward with sequencing n=3 from each time point in the ambient. I need to miRNA library prep 24 more samples. Today, I re-amped samples M8, M14, M28, M39, M48, M51, M61, M63, M74, M75, M87, and M88. M8, M14, M28, M48, and M51 were extracted on 2/22/24 and M39, M61, M63, M74, M75, M87, and M88 were extracted on 5/2/24.
I received new TaqPreMix from Zymo so I redid sections 4, 5, and QC on 7/24/24.
Here’s the miRNA library prep workflow:
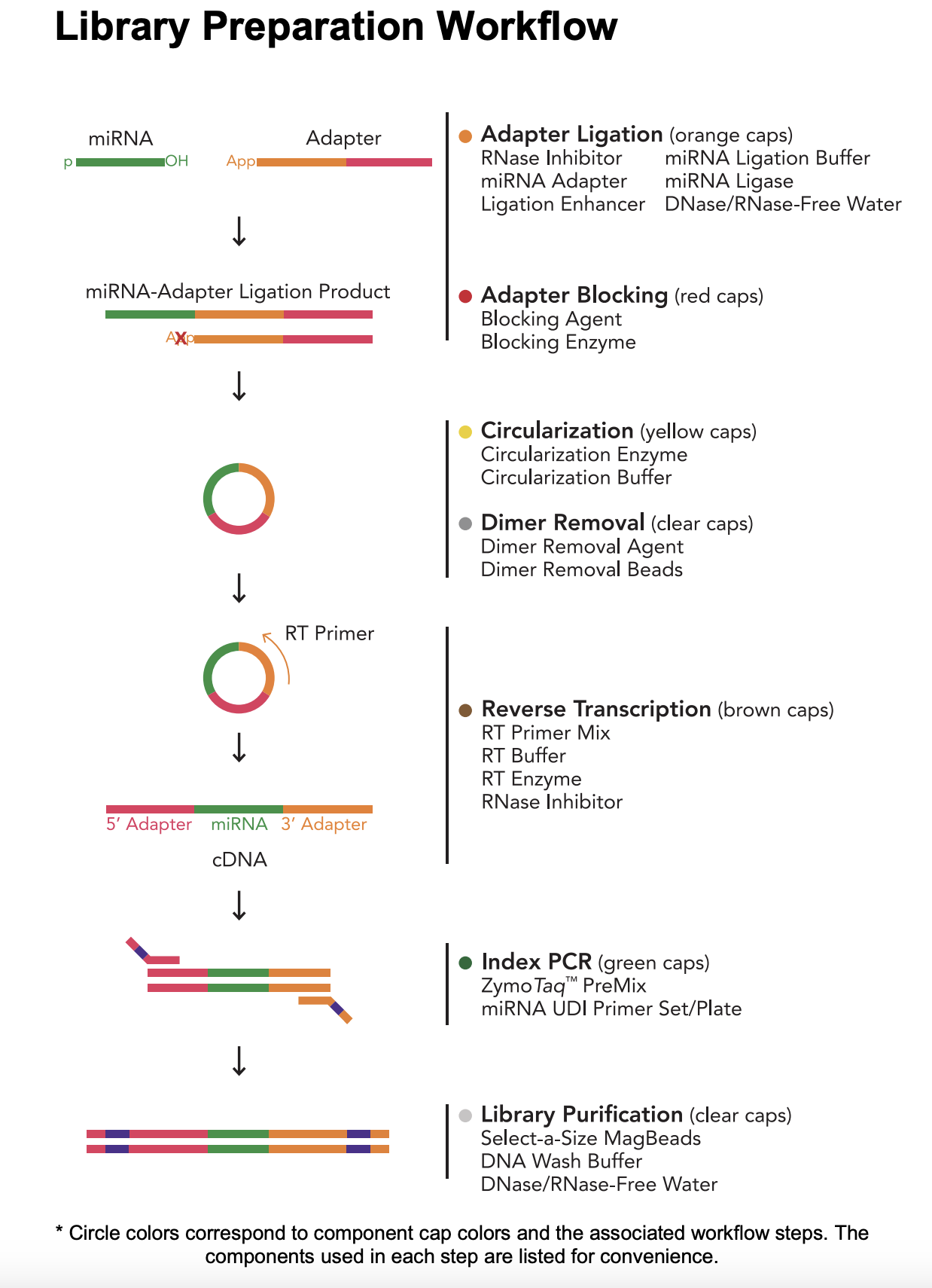
Materials
- Zymo-Seq miRNA library kit
- PCR tubes
- Thermocycler
- Heating block
- Mini centrifuge
- Bench top centrifuge
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- Kit contents
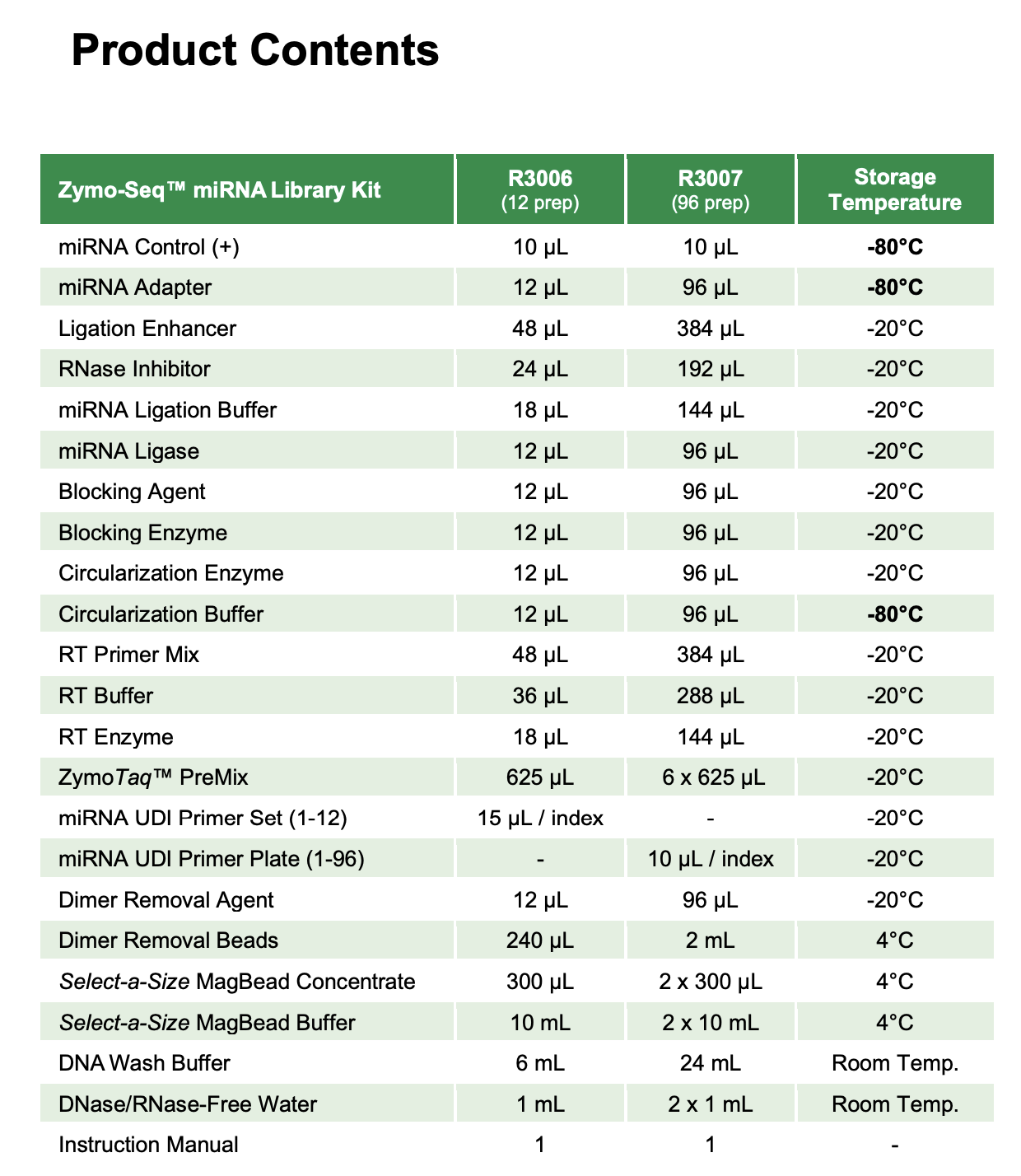
Materials
Protocol was followed according to this post. A few differences:
- I received new TaqPreMix from Zymo so I redid sections 4, 5, and QC on 7/24/24.
QC
Run DNA Tapestation for visualize libraries. Here’s an example of what the library should look like on a Tapestation:
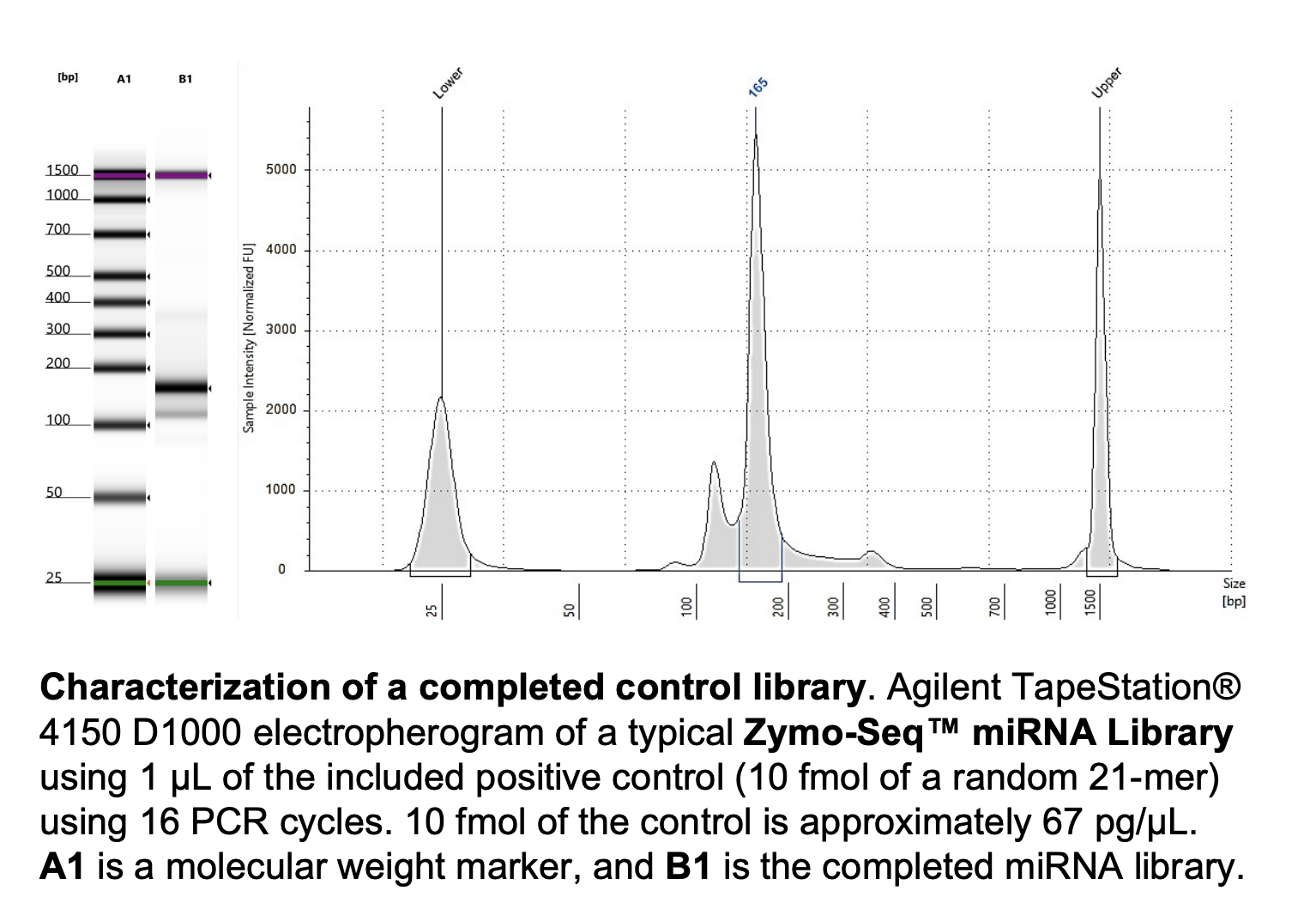
See full sample report here.
Much better!!! New Taq mix was the way to go. Some samples have two peaks so I think I will do a bead cleanup before sending.