SwitchFree library prep for ENCORE priming
SwitchFree library prep
This post details the info about the SwitchFree library prep. See Zymo’s protocol here.
I will be prepping samples from Flo’s ENCORE priming experiment conducted in Bermuda 2024 (see github repo for more information on the experiment). Today, I am prepping 8 samples.
The kit needs a minimum of 10 ng of total RNA or a maximum of 500 ng of total RNA, which is a large range. We will be using 12 ng of total RNA as input. Here’s a breakdown of input RNA volumes for each sample:
| TubeID | Qubit RNA avg (ng/uL) | Strip tube # | RNA (uL) | Ultrapure water (uL) | Total starting volume (ul) | Primer |
|---|---|---|---|---|---|---|
| MD-4-21 | 15.2 | 1 | 1 | 4 | 5.0 | 1 |
| MD-5-6 | 15.1 | 2 | 1 | 4 | 5.0 | 2 |
| MD-2-23 | 10.15 | 3 | 1.2 | 3.8 | 5.0 | 7 |
| MD-2-8 | 13.6 | 4 | 1 | 4 | 5.0 | 58 |
| MD-1-4 | 13.7 | 5 | 1 | 4 | 5.0 | 85 |
| MD-5-20 | 4.79 | 6 | 2.5 | 2.5 | 5.0 | 86 |
| MD-5-3 | 43.5 | 7 | 1 | 4 | 5.0 | 87 |
| MD-1-7 | 27.6 | 8 | 1 | 4 | 5.0 | 88 |
Here’s the SwitchFree library prep workflow:
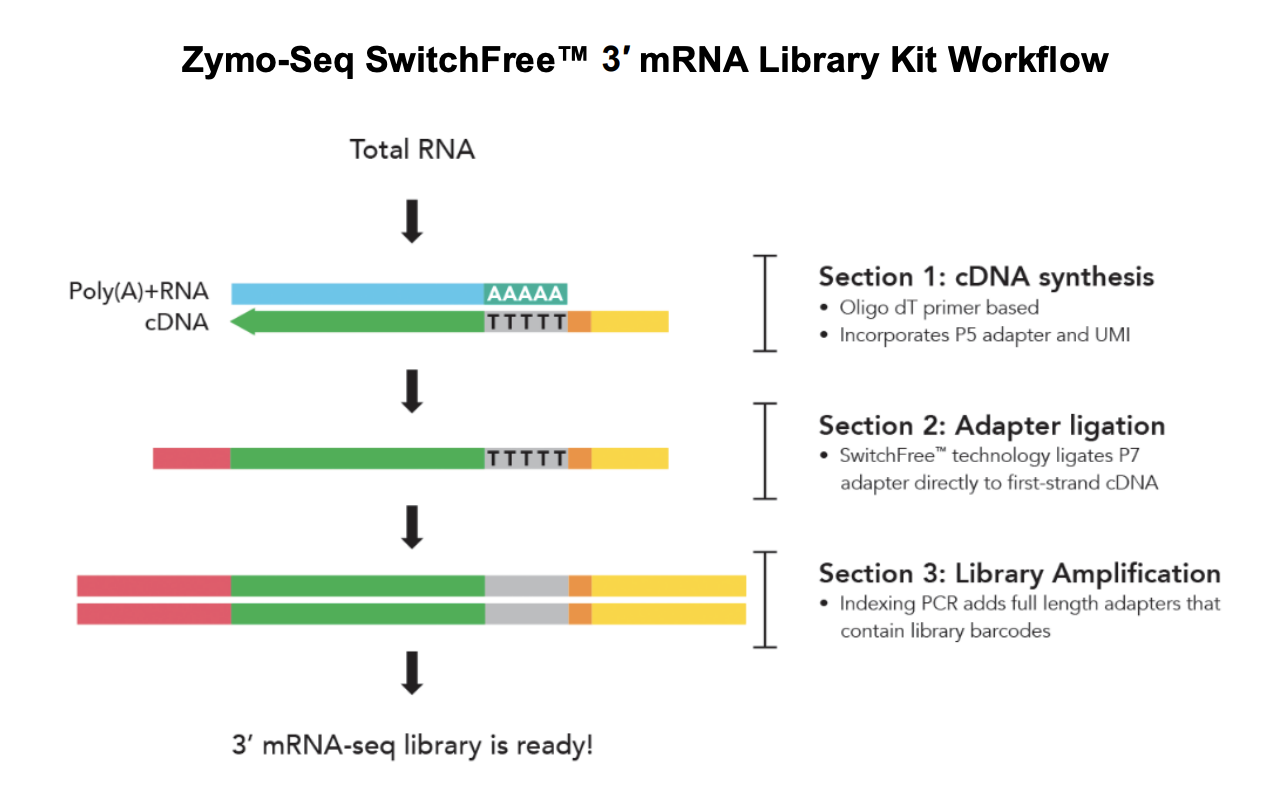
Materials
- Zymo-Seq SwitchFree 3’ mRNA Library Kit
- PCR tubes
- Thermocycler
- Mini centrifuge
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- Kit contents
Buffer preperation
Once buffers are prepared for a kit, they do not need to be prepared again.
- Add 300 uL of the Select-a-Size MagBead concentrate to 10 mL of Select-a-Size MagBead Buffer.
- These should be prepared at least 5 days ahead of library prep.
- Add 24 mL of 100% ethanol to the DNA Wash Buffer concentrate. Store at room temperature.
Best practices
- Avoid multiple freeze thaws of all components, and aliquot components as necessary
- Remove enzymes from cold storage just before use and return to cold storage immediately after use
- Thaw and maintain all components on ice unless noted otherwise
- Flick to mix thawed components and briefly centrifuge prior to use
- After adding each component, mix by pipetting up and down 15-20 times. Briefly centrifuge after
- Pre-program thermal cycler with lid heating ON set to >100-105°C
- Turn on thermocycler day-of to preheat
- Pre-calculate how much to dilute RNA samples
Protocol
Protocol was followed according to this post.
Some modifications
- Used 12 ng RNA input instead of 11 ng
- For the polyA R1 reagent, used 3uL of polyA R1 reagent + 2uL of DNase/RNase free water instead of 5 uL of polyA R1 reagent
- Sections 1 and 3 were done on 6/30/25; Section 3 and QC were done on 7/1/25
- In Section 3, after the thermocycler library amplification, samples sat at 4C for ~45 mins while I had a brief meeting
QC
Run DNA Tapestation to visualize libraries. Here’s an example of what the library should look like on a Tapestation:
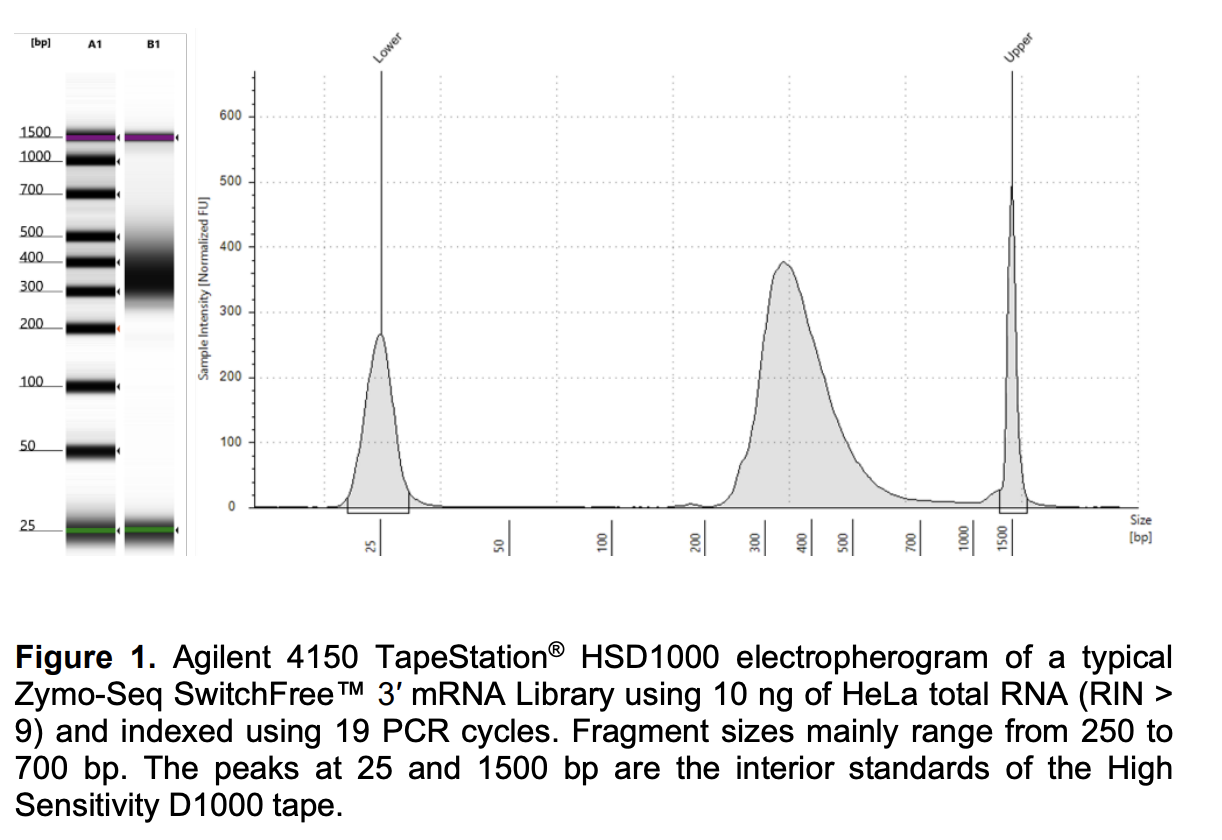
I ran the tapestation on the libraries on 7/1/25. See the full report here.








