SwitchFree library prep
SwitchFree library prep test
This post details the info about the SwitchFree library prep steps. Hollie got this kit as a test to see if we can produce adequate libraries for RNA sequencing with different species. I’m using the Zymo-Seq SwitchFree 3’ mRNA Library Kit. See Zymo’s protocol here.
We will be using samples 8, 9, 106, 109, 111, and 112 from the POC 2023 spawning project. These samples were extracted on 4/3/24. The kit needs a minimum of 10 ng of total RNA or a maximum of 500 ng of total RNA, which is a large range. Since some of the concentrations in these samples were low, we will be using 11 ng of total RNA as input. Here’s a breakdown of input RNA volumes for each sample:
| TubeID | TS RNA (ng/uL) | Strip tube # | RNA (uL) | Ultrapure water (uL) | Total starting volume (ul) | Primer |
|---|---|---|---|---|---|---|
| 8 | 15.9 | 1 | 0.7 | 4.3 | 5.0 | 7 |
| 9 | 23.2 | 2 | 0.5 | 4.5 | 5.0 | 8 |
| 106 | 7.29 | 3 | 1.5 | 3.5 | 5.0 | 9 |
| 109 | 8.71 | 4 | 1.3 | 3.7 | 5.0 | 10 |
| 111 | 7.23 | 5 | 1.5 | 3.5 | 5.0 | 11 |
| 112 | 6.27 | 6 | 1.8 | 3.2 | 5.0 | 12 |
Here’s the SwitchFree library prep workflow:
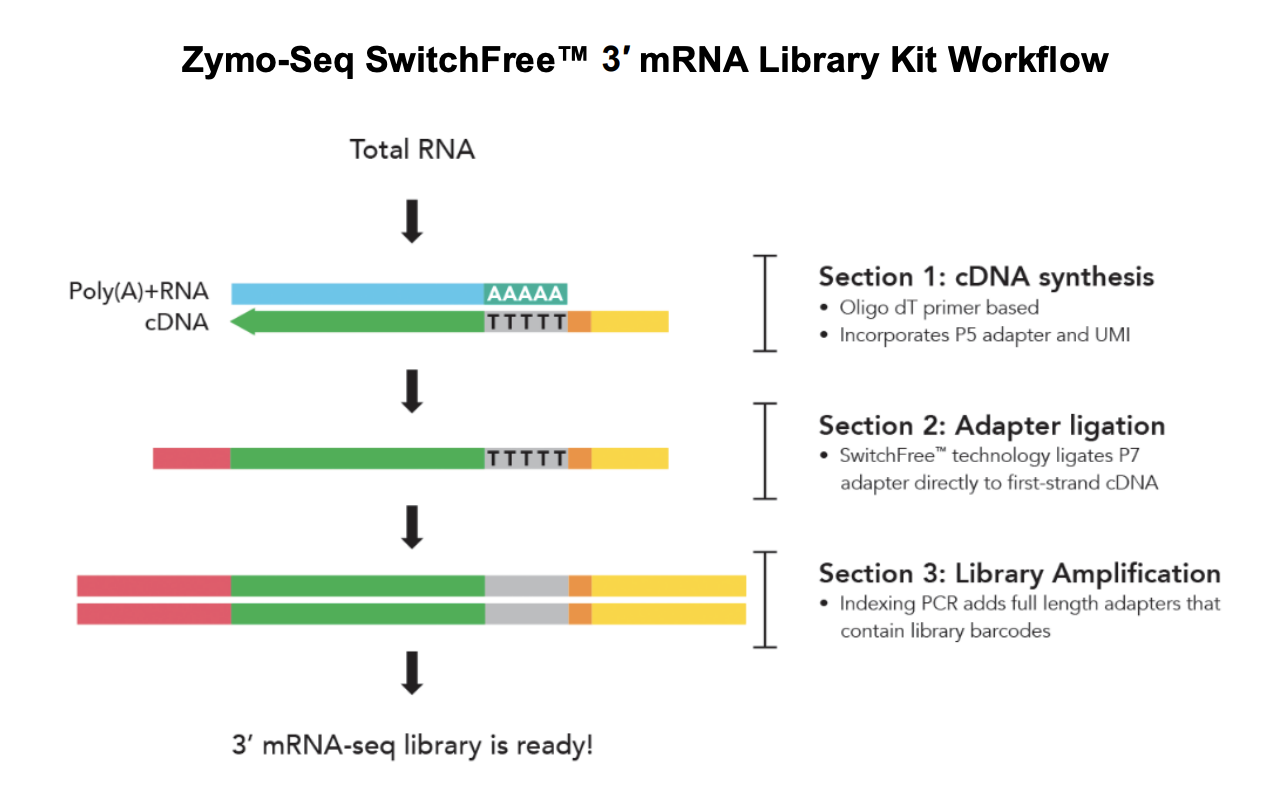
Materials
- Zymo-Seq SwitchFree 3’ mRNA Library Kit
- PCR tubes
- Thermocycler
- Mini centrifuge
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- Kit contents
Buffer preperation
Once buffers are prepared for a kit, they do not need to be prepared again.
- Add 300 uL of the Select-a-Size MagBead concentrate to 10 mL of Select-a-Size MagBead Buffer.
- These should be prepared at least 5 days ahead of library prep.
- Add 24 mL of 100% ethanol to the DNA Wash Buffer concentrate. Store at room temperature.
Best practices
- Avoid multiple freeze thaws of all components, and aliquot components as necessary
- Remove enzymes from cold storage just before use and return to cold storage immediately after use
- Thaw and maintain all components on ice unless noted otherwise
- Flick to mix thawed components and briefly centrifuge prior to use
- After adding each component, mix by pipetting up and down 15-20 times. Briefly centrifuge after
- Pre-program thermal cycler with lid heating ON set to >100-105°C
- Turn on thermocycler day-of to preheat
- Pre-calculate how much to dilute RNA samples
Protocol
Protocol was followed according to this post.
Sections 1 and 2 were done on 4/4/24. Section 3 was done on 4/5/24.
QC
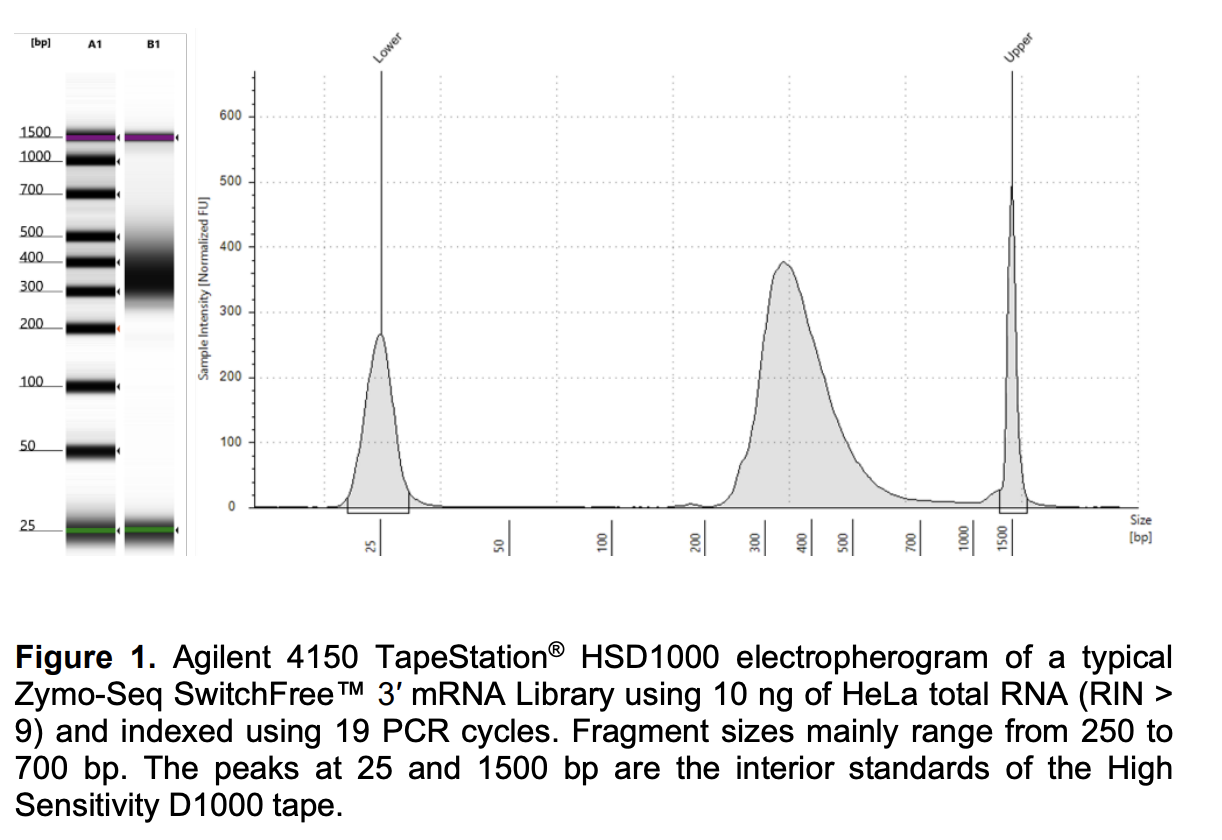
Run DNA Tapestation to visualize libraries. Here’s an example of what the library should look like on a Tapestation:
I ran the tapestation on the libraries on 4/5/24. See the full report here.
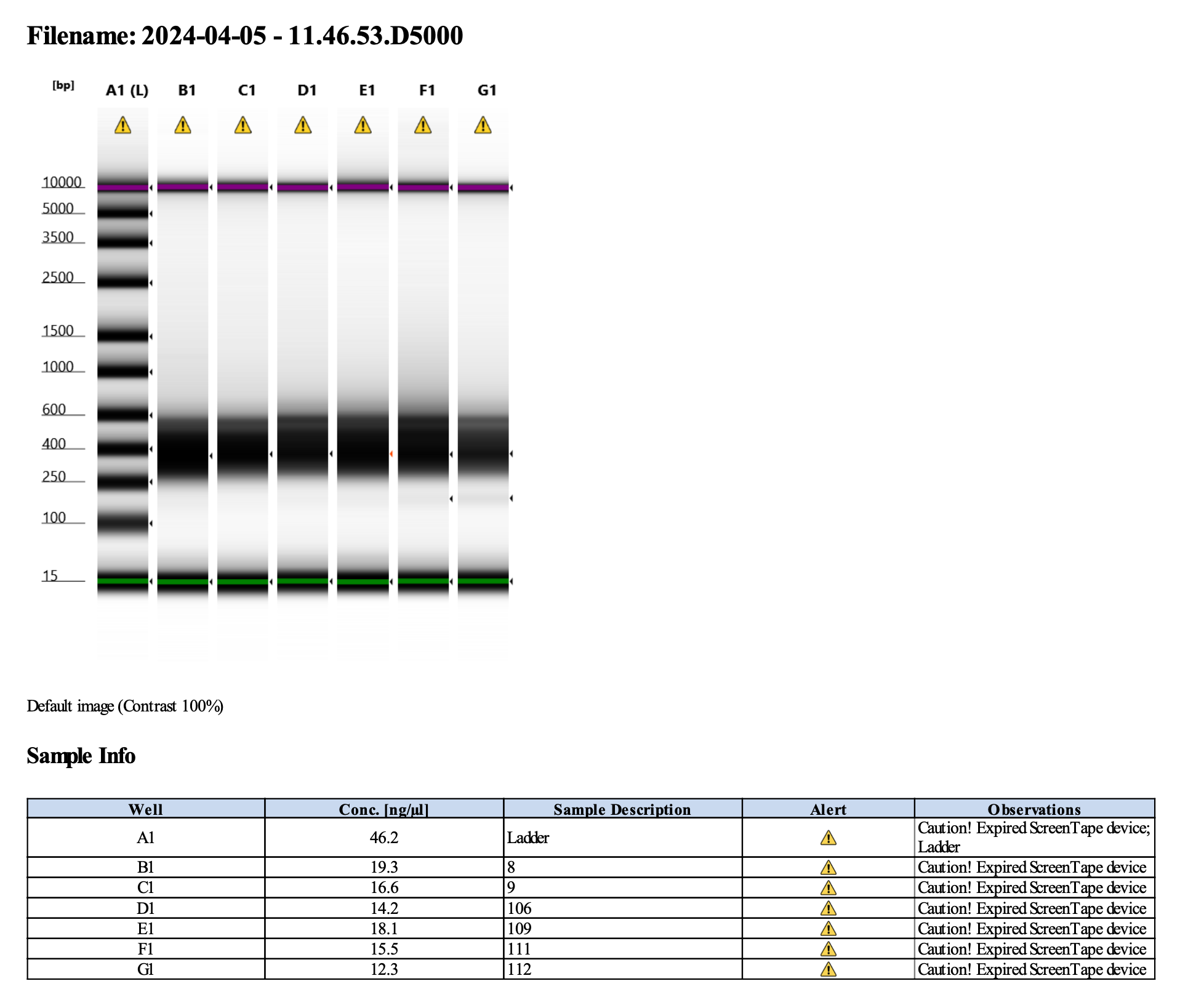
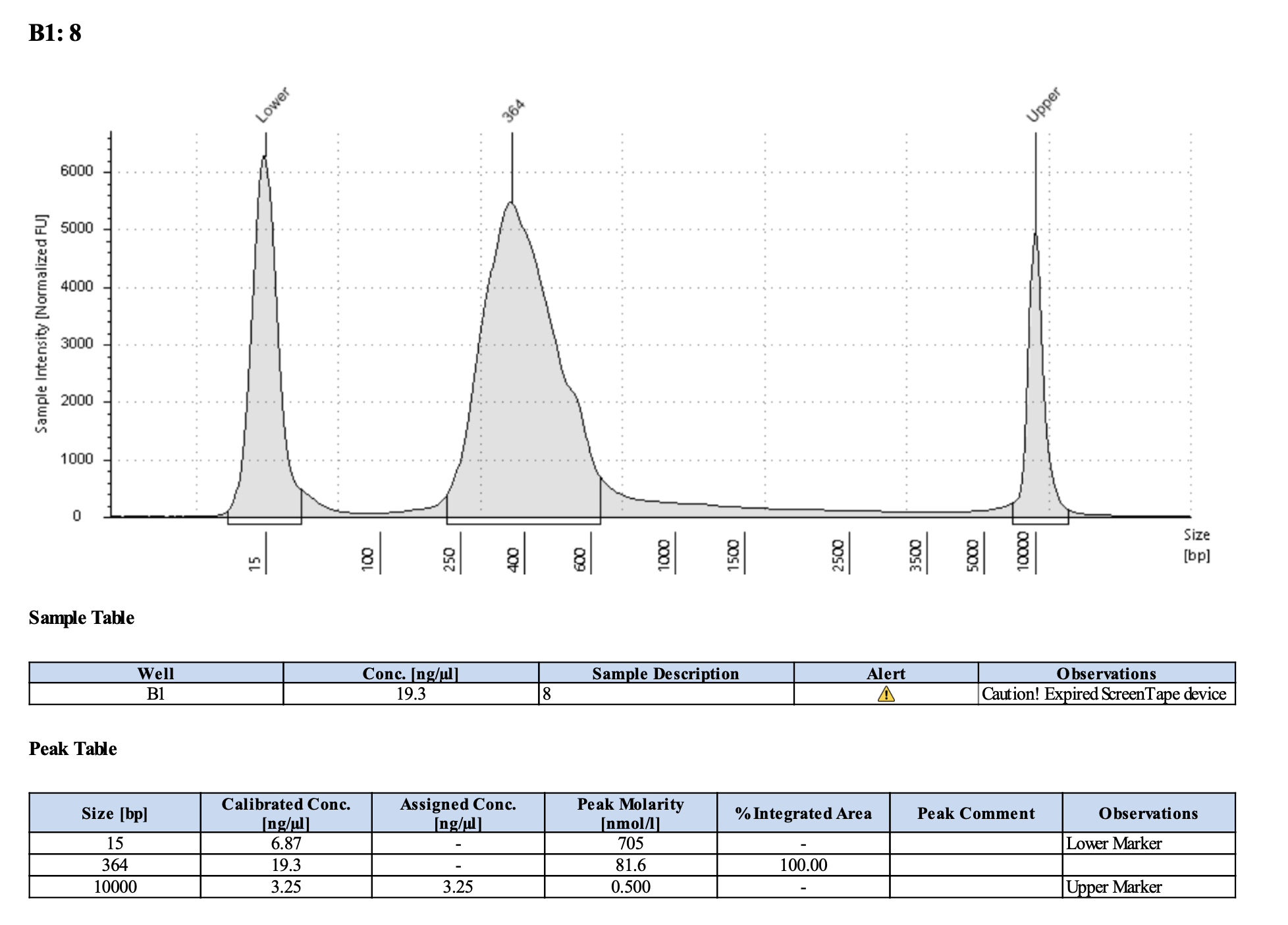
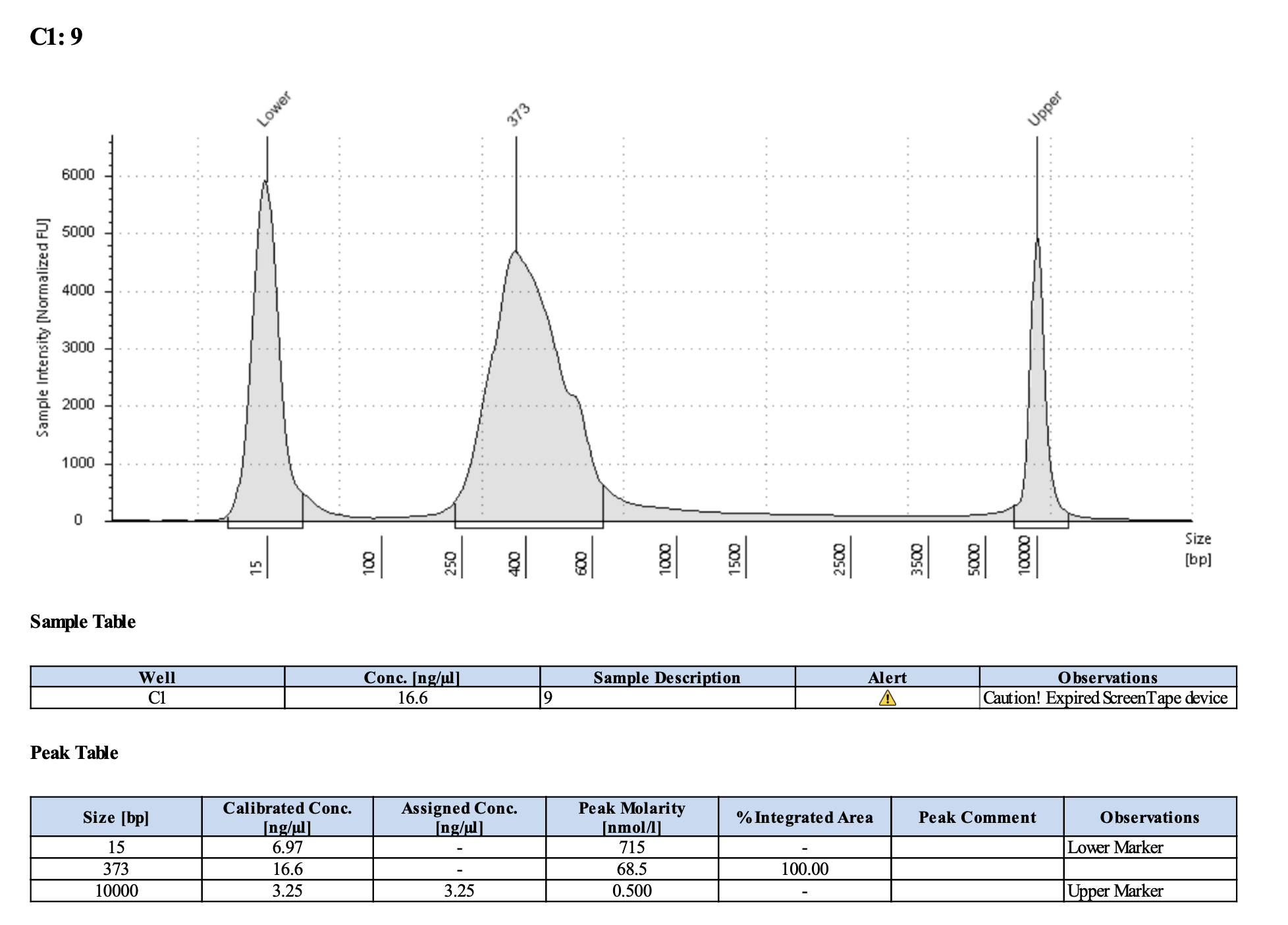
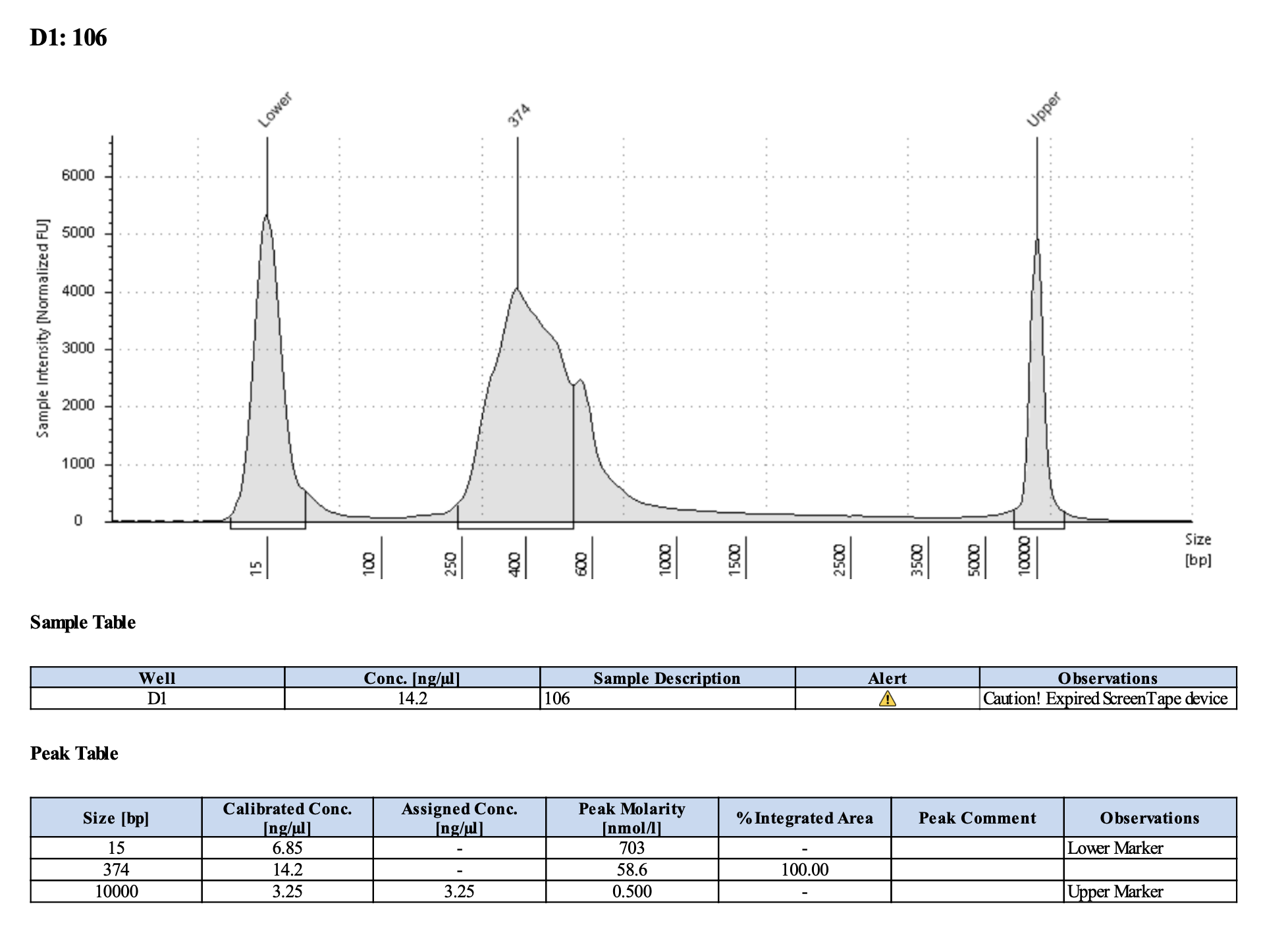
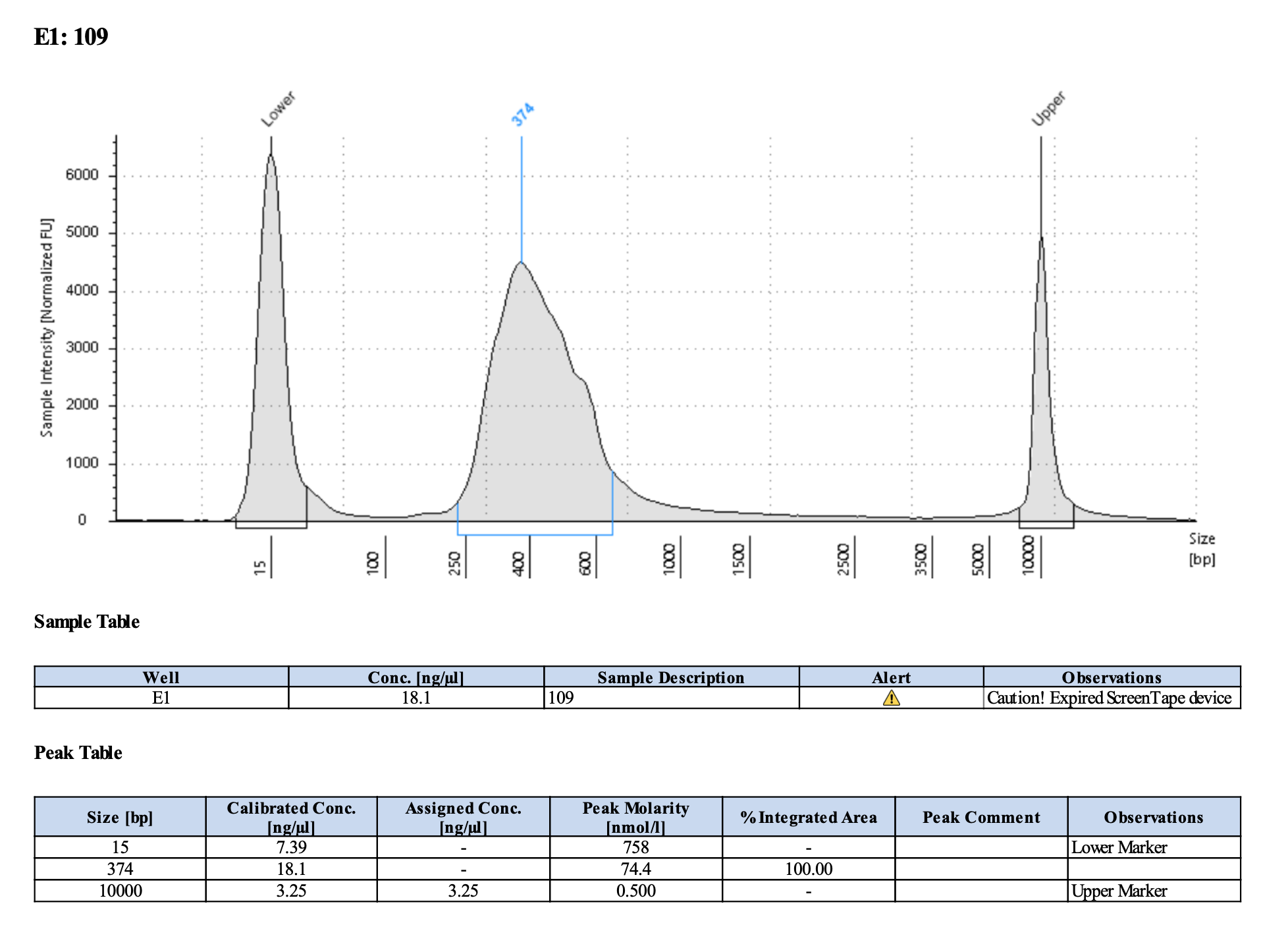
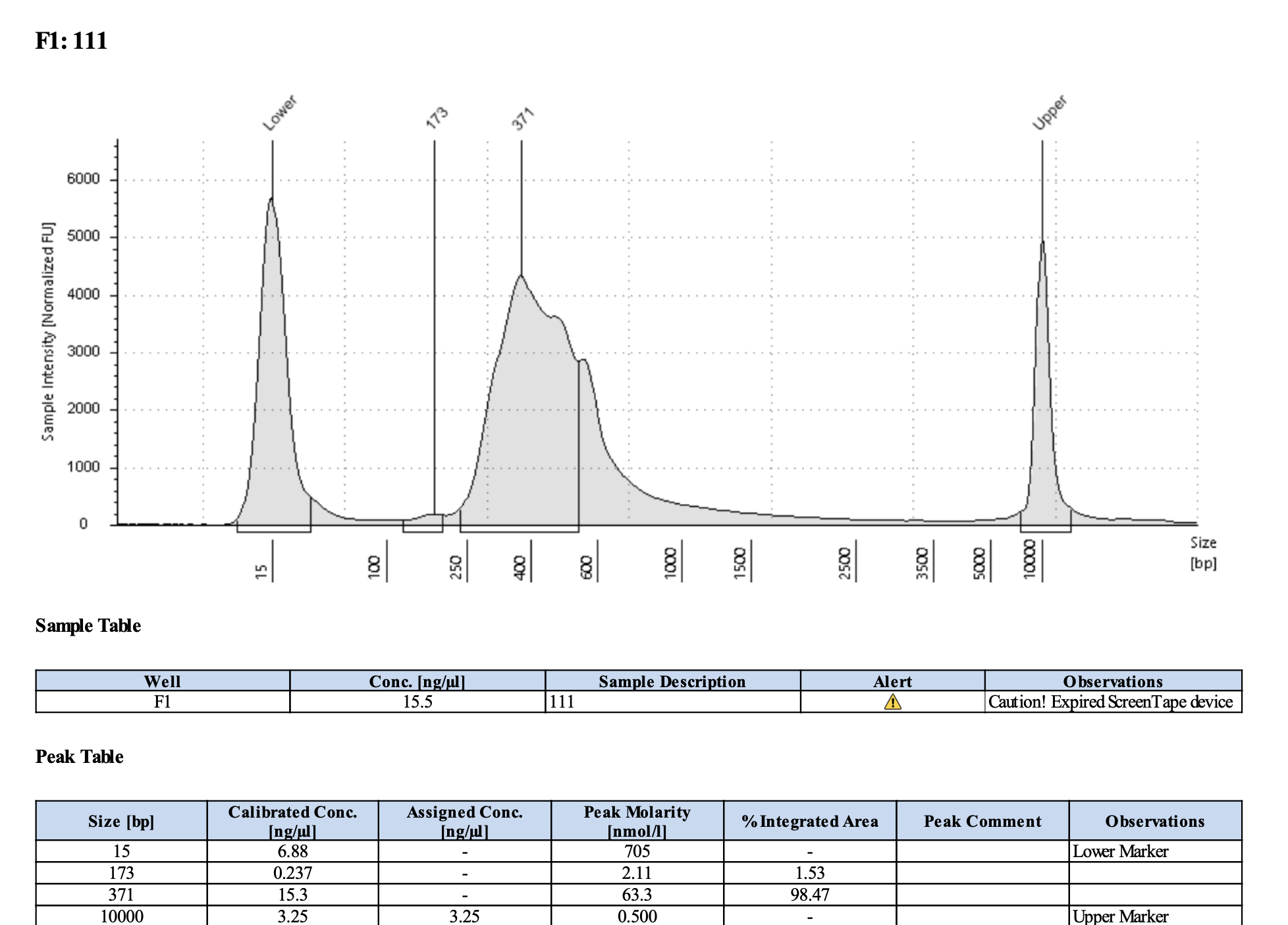
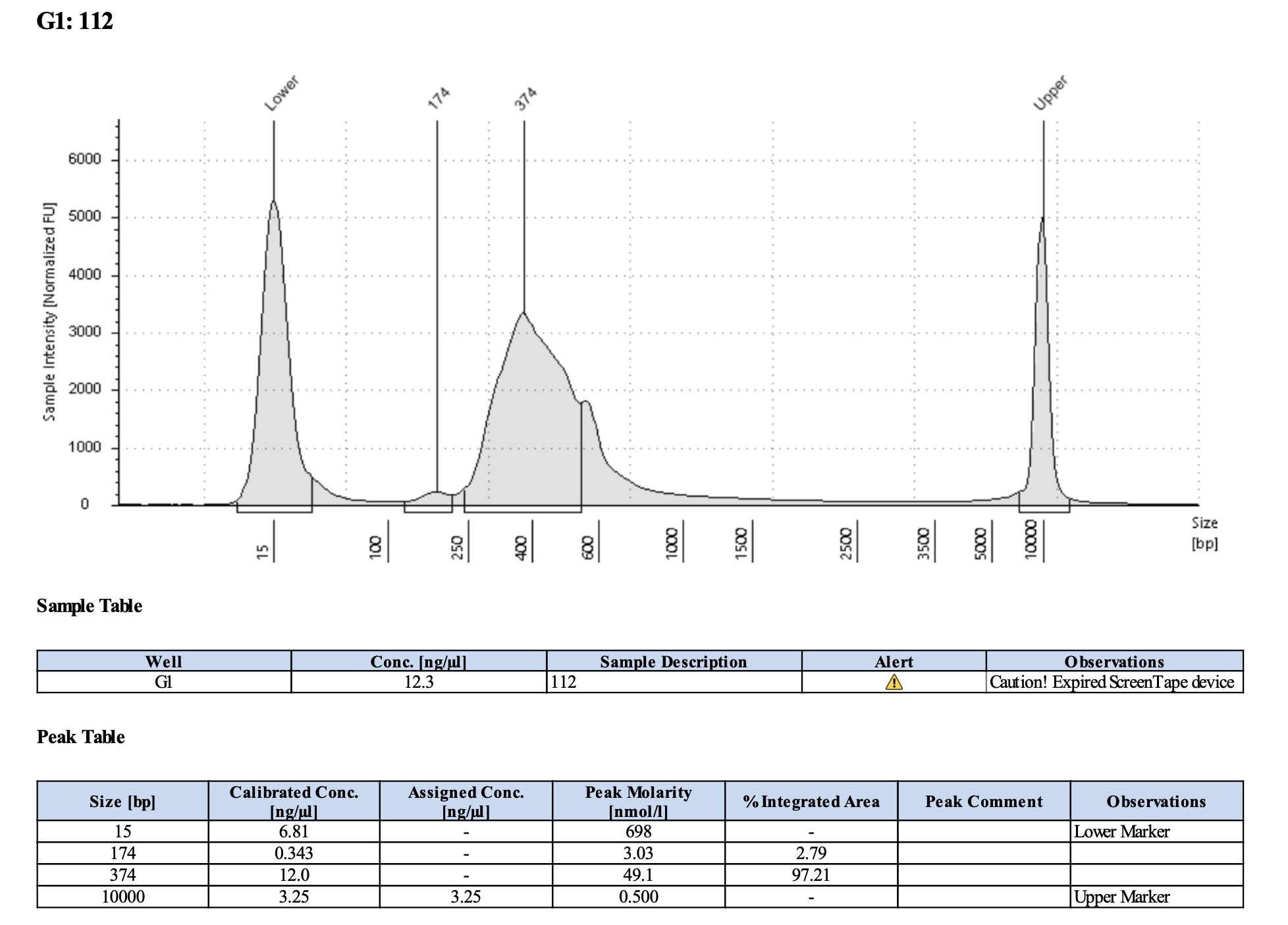
Tapestation peaks and concentrations look great for all samples! All of the samples have a slight shoulder to the right, which might indicate PCR bubbles (read more here). Libraries with a bubble product can still be sequenced, which is good. I discussed with Hollie and we will move forward with sequencing. I will provide Genohub with the tapestation results, as well as the Zymo manual.