RiboFree library prep
RiboFree library prep test for Mcap developmental time series Hawaii 2023
This post details the info about the library prep steps for the M. capitata developmental time series experiment in Hawaii 2023. The github for that project is linked here. I’m using the Zymo-Seq RiboFree Total RNA Library Kit for total RNA library prep (I got the 12 prep kit). See Zymo’s protocol here.
I’ll be using samples M9, M13, M23, M35, M52 and M85. M9 was extracted on 2/22/24, and all of the other samples were extracted on 2/10/24. These samples, along with M60 and M72 (prepped on 2/19/24), will be sent for sequencing. The samples represent n=1 from each time point during M. capitata development. The kit needs a minimum input of 10 of total RNA or a maximum input of 250 ng of total RNA. I’m going to use 190 ng as my input concentration; Maggie did a similar input for her ribofree preps). Here’s the breakdown of input RNA volumes for each sample:
| TubeID | Qubit RNA Average (ng/uL) | Strip tube # | RNA (uL) | DNA/RNA free water (uL) | Total starting volume (uL) | Primer |
|---|---|---|---|---|---|---|
| M9 | 16.6 | 1 | 8 | 0 | 8 | 5 |
| M13 | 58.5 | 2 | 3.5 | 4.5 | 8 | 6 |
| M23 | 15.4 | 3 | 8 | 0 | 8 | 7 |
| M35 | 29 | 4 | 6.5 | 1.5 | 8 | 8 |
| M52 | 19 | 5 | 8 | 0 | 8 | 9 |
| M85 | 30.3 | 6 | 6.5 | 1.5 | 8 | 10 |
Here’s the RiboFree library prep workflow:
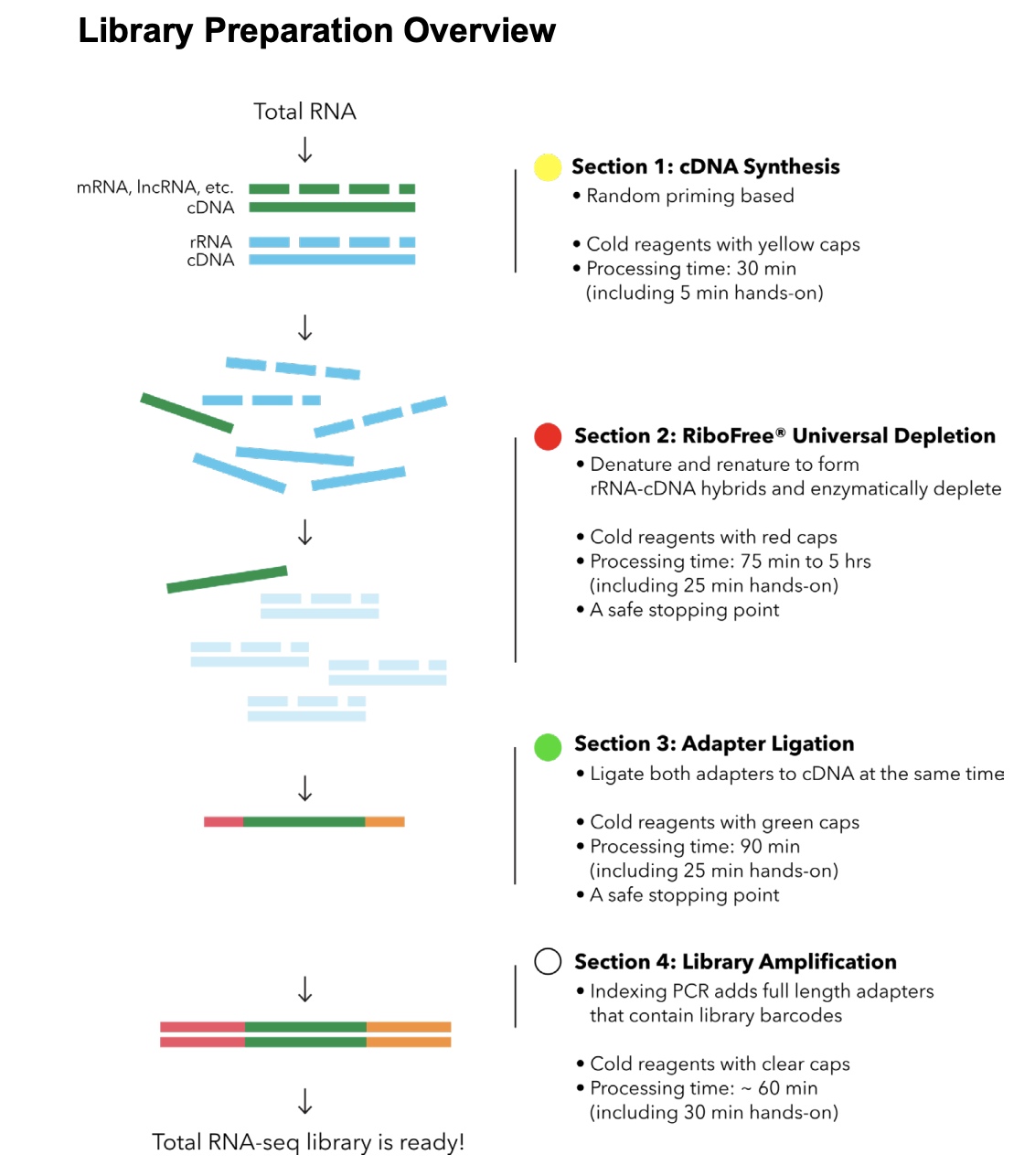
Materials
- Zymo-Seq RiboFree library kit
- PCR tubes
- Thermocycler
- Heating block
- Mini centrifuge
- Vortex
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- 1.5 mL tubes
- Kit contents
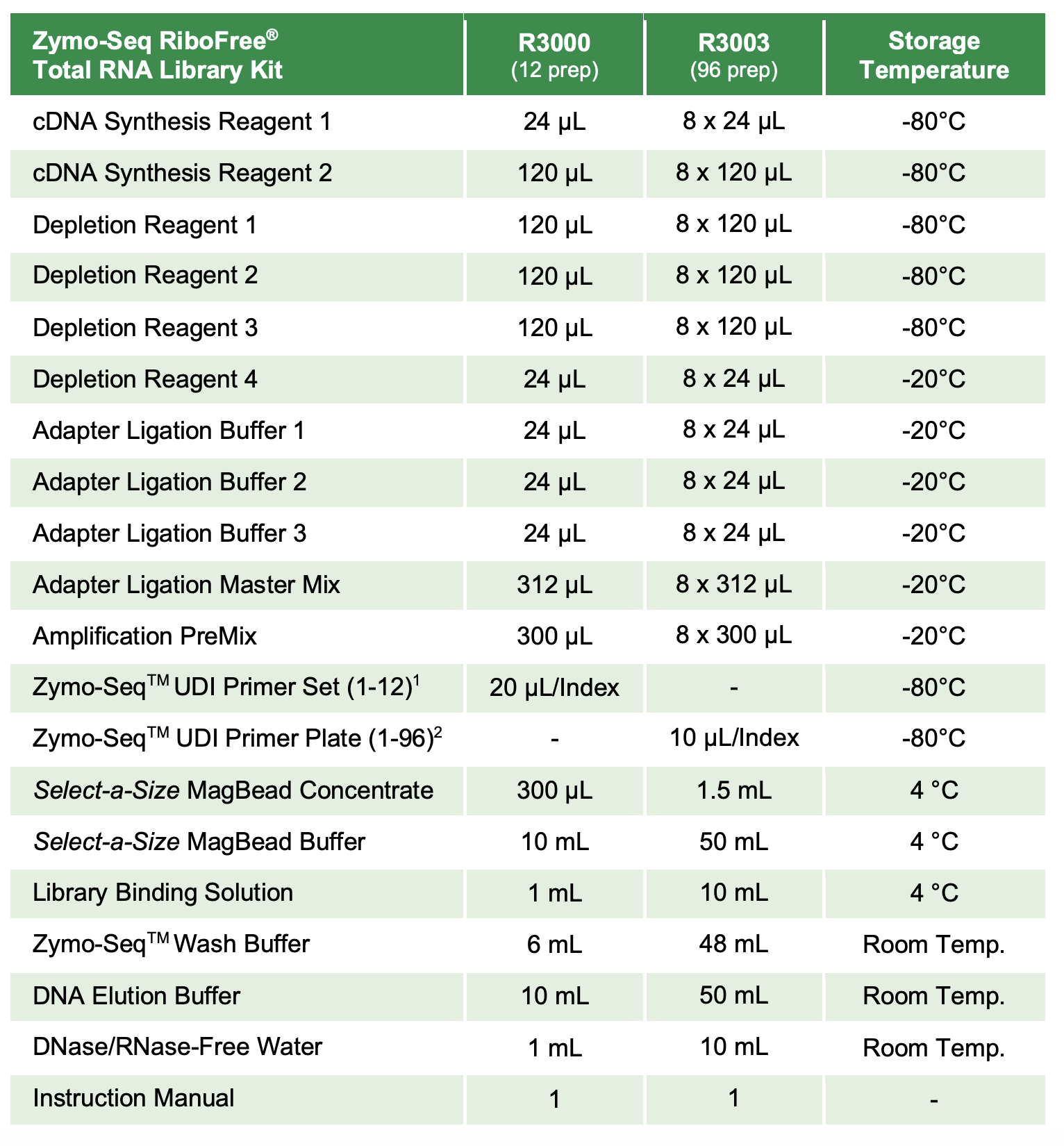
Protocol
Protocol was followed according to this post. The only difference is that the number of PCR library amplification cycles was changed to 15 from 11. M85 also got ~8 uL of Depletion Reagent 1 instead of 10 uL due to a pipetting error. See table above for which primer number was used for each sample.
QC
Run DNA Tapestation for visualize libraries. Here’s an example of what the library should look like on a Tapestation:
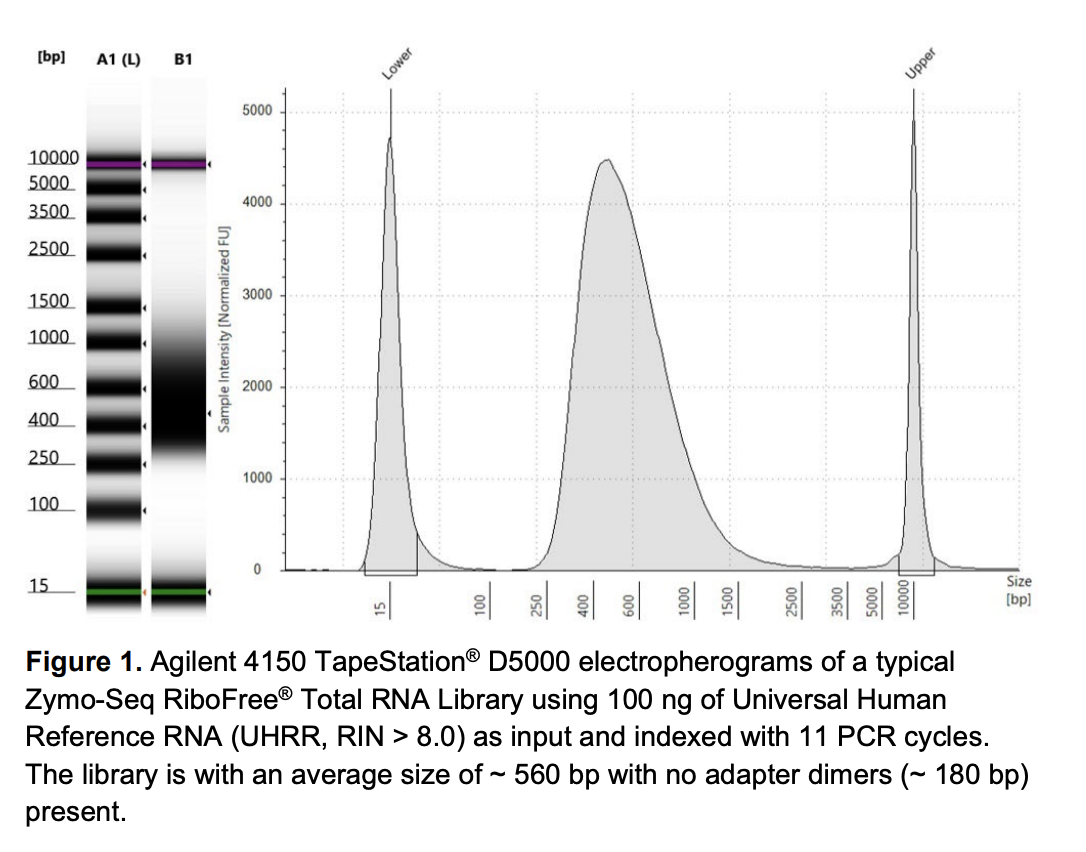
Here’s how my samples turned out on 2/26/24. See full report here.
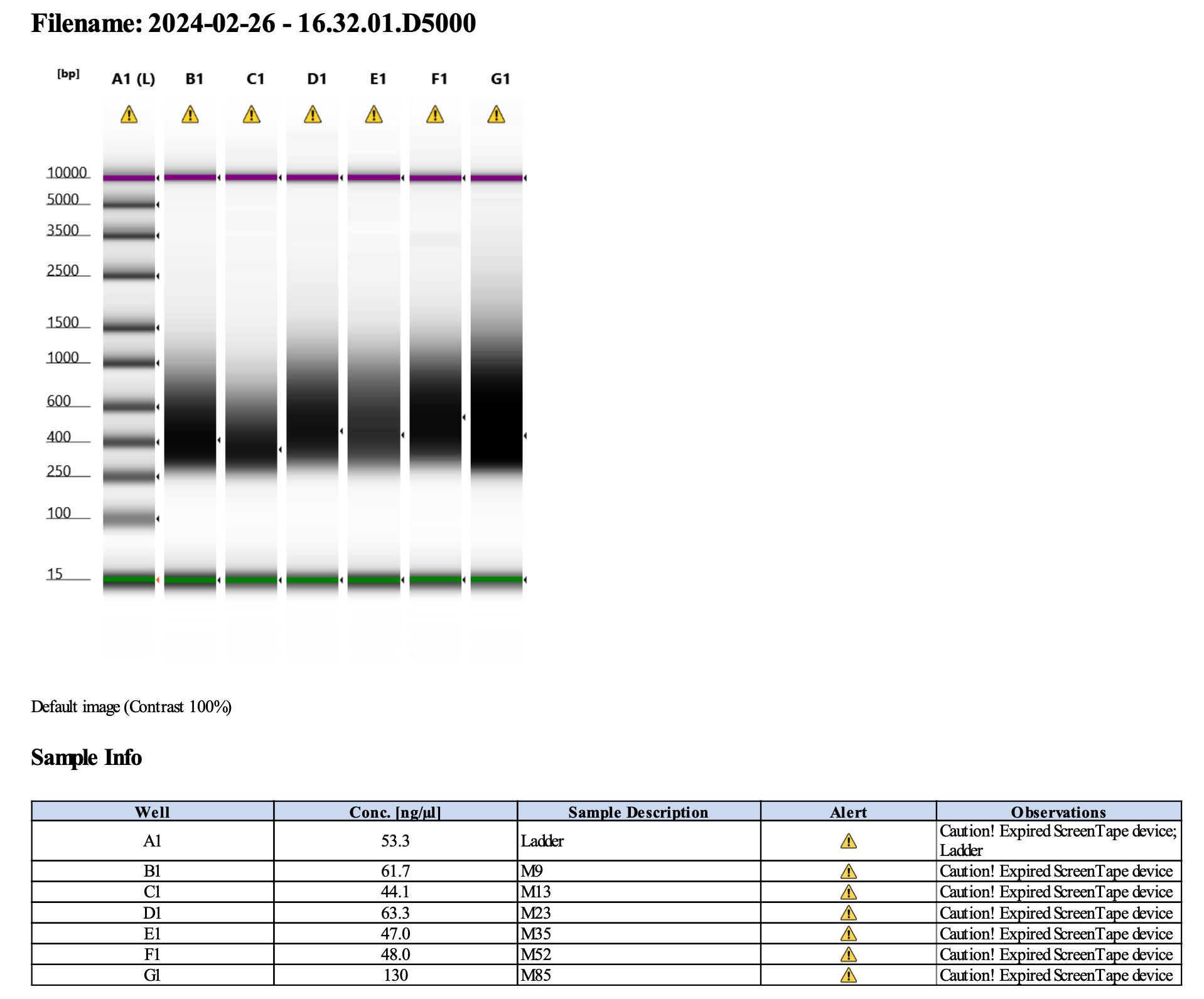
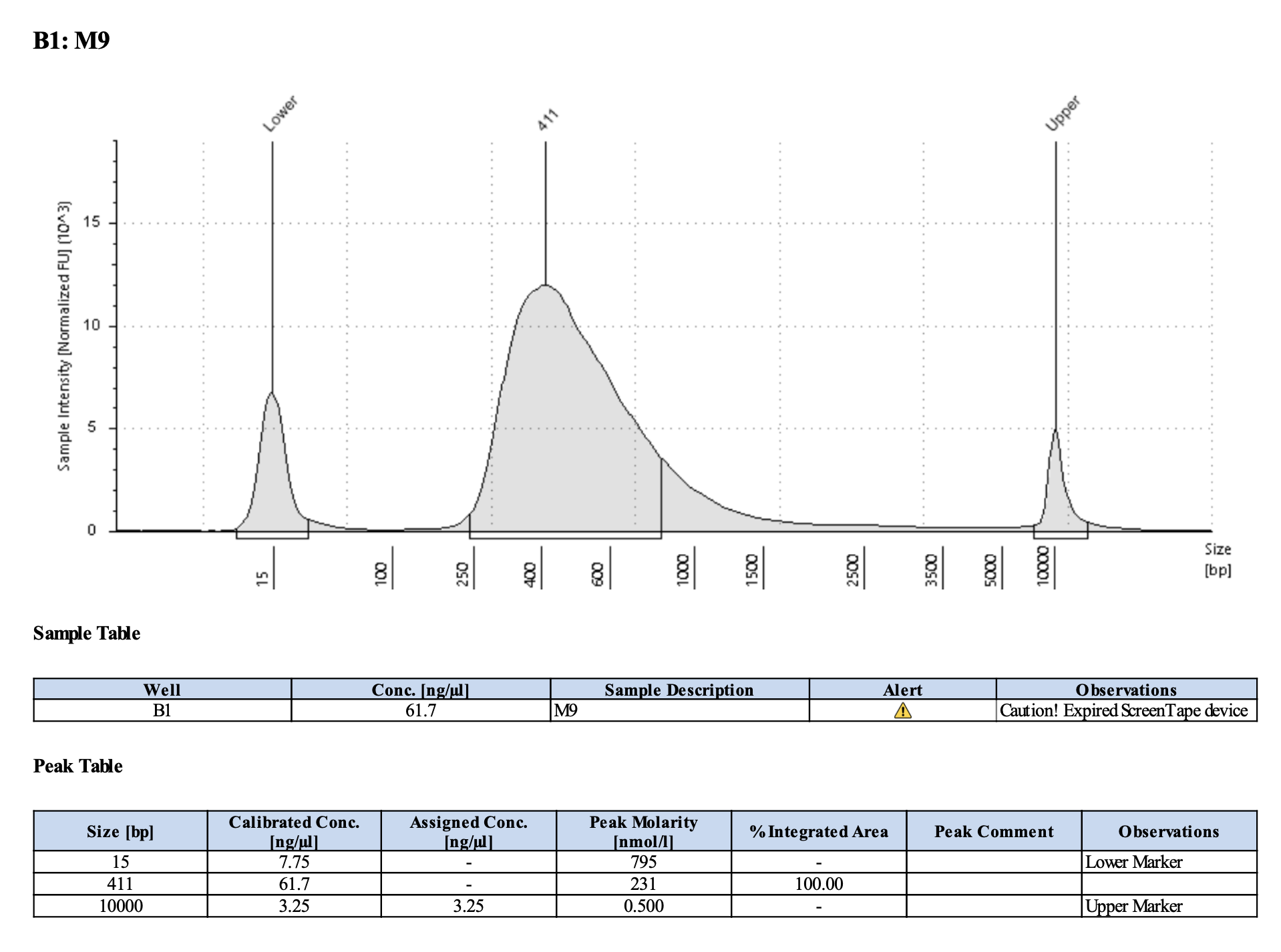
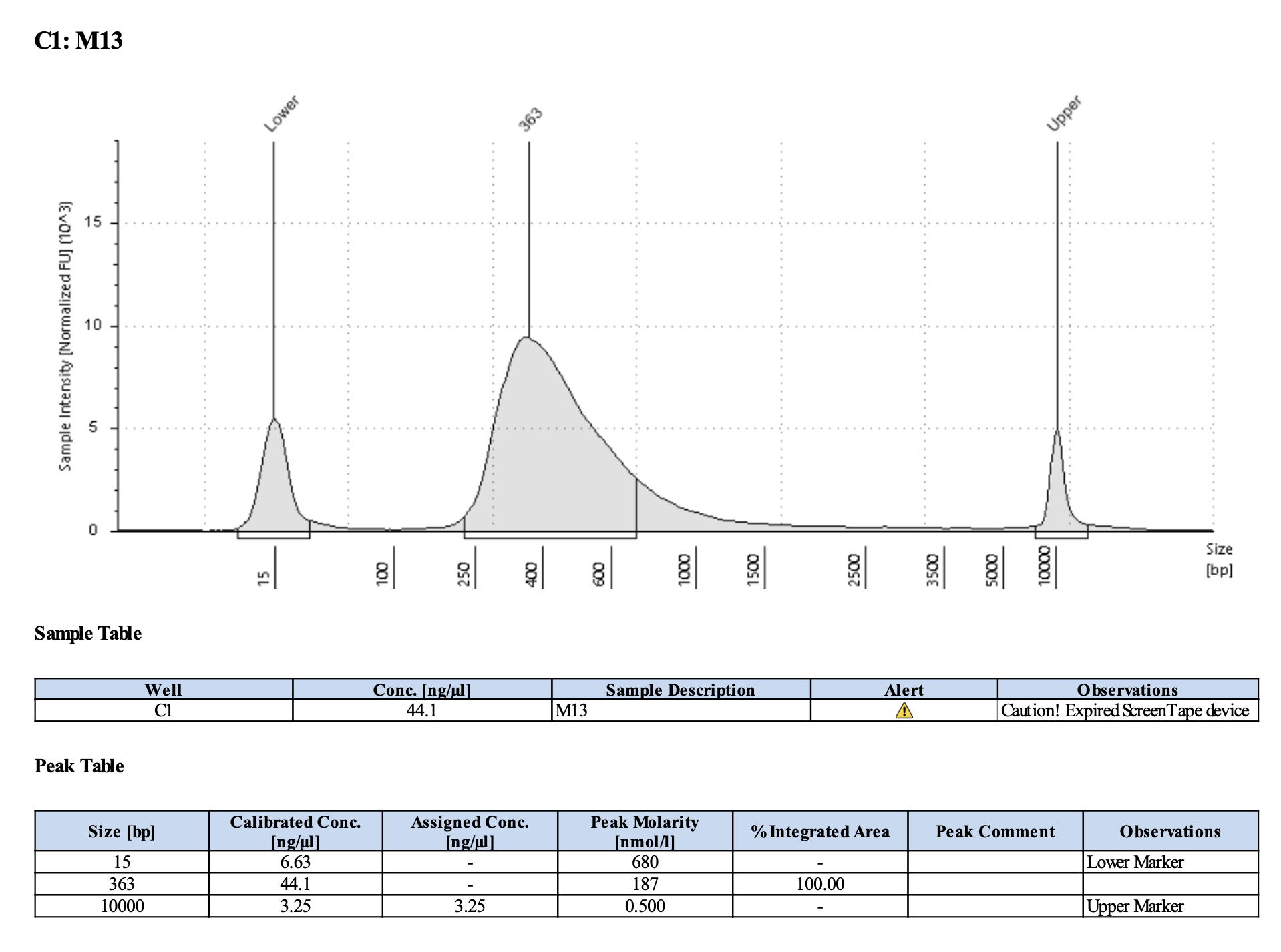
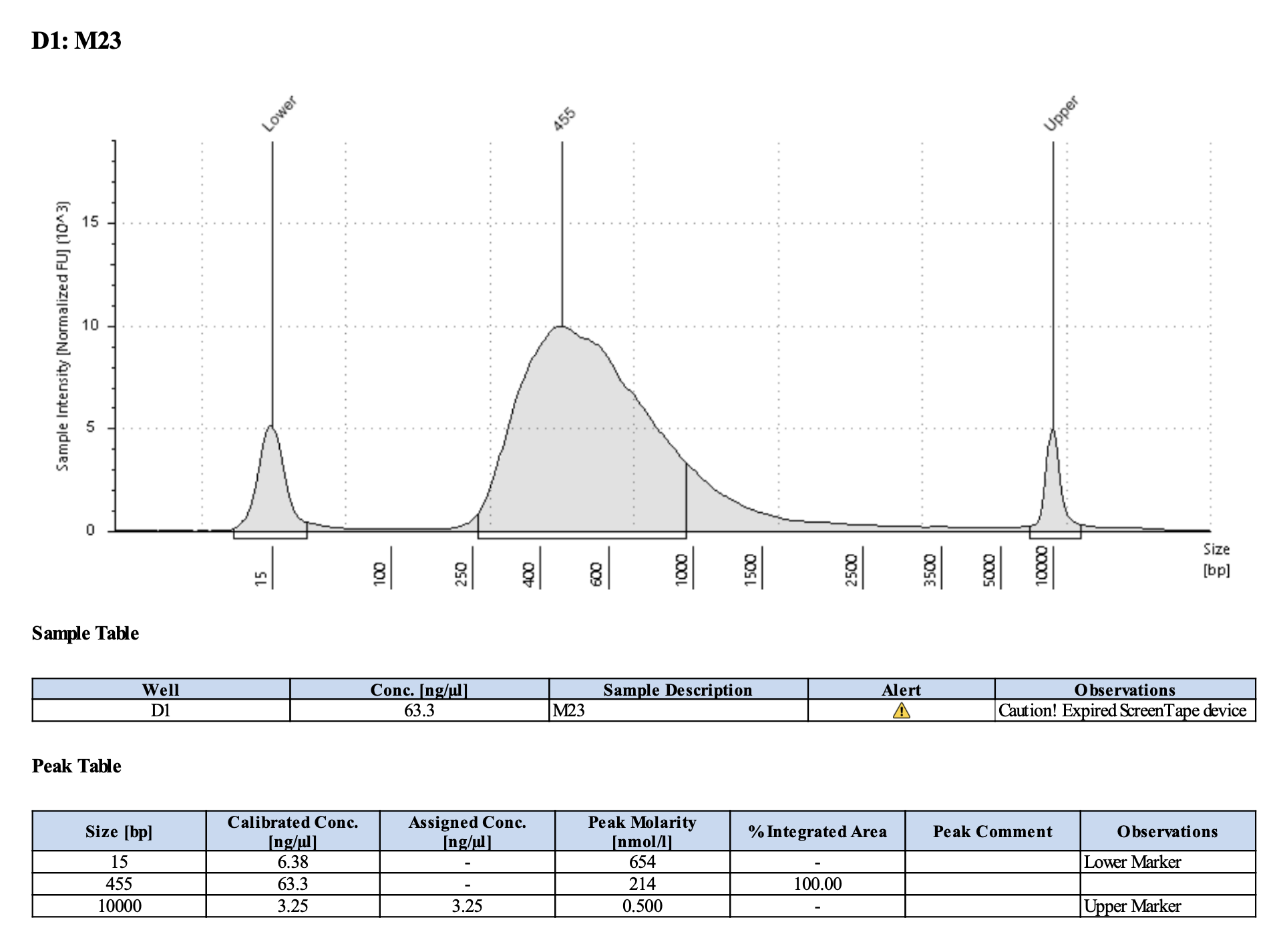
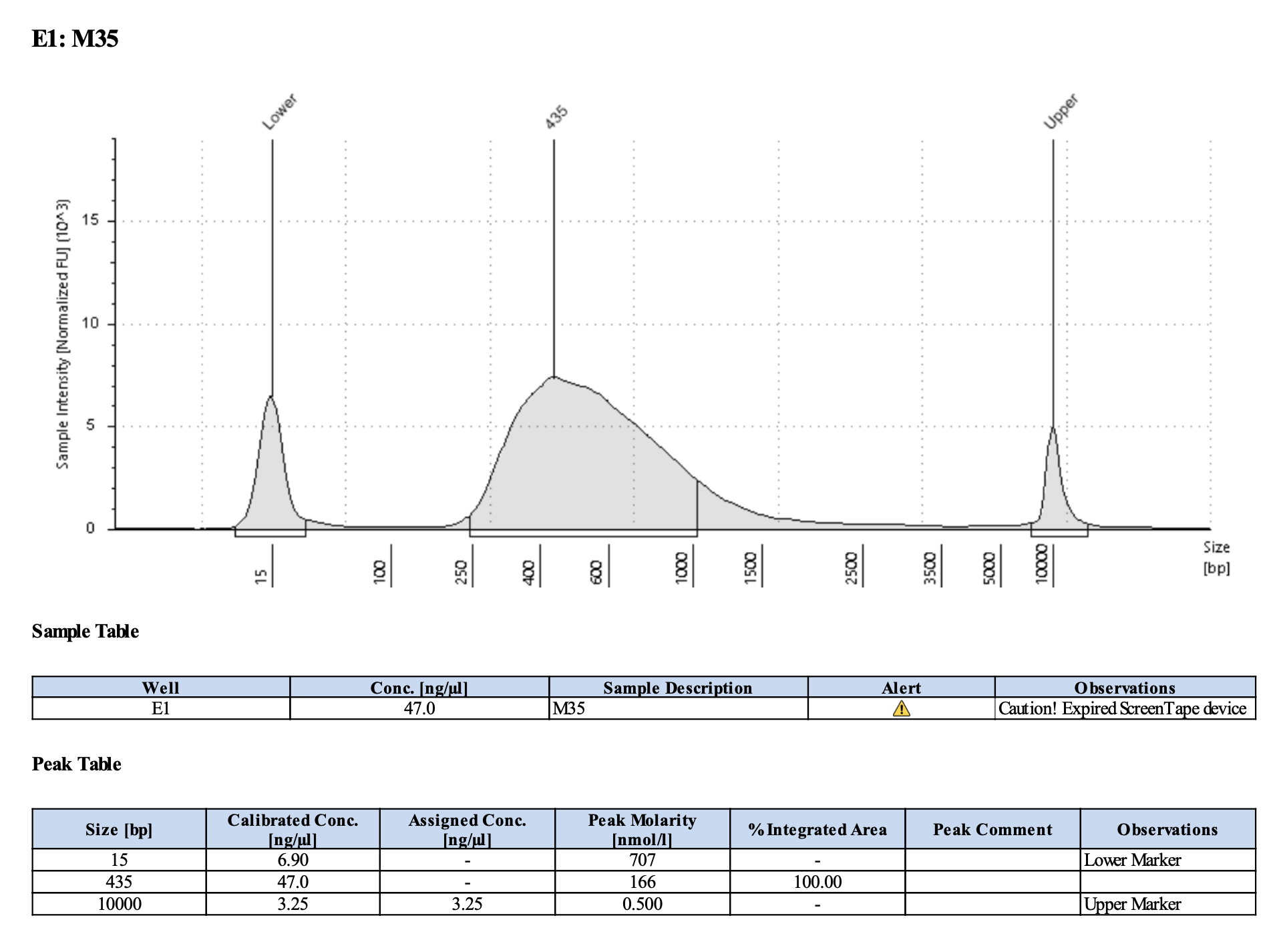
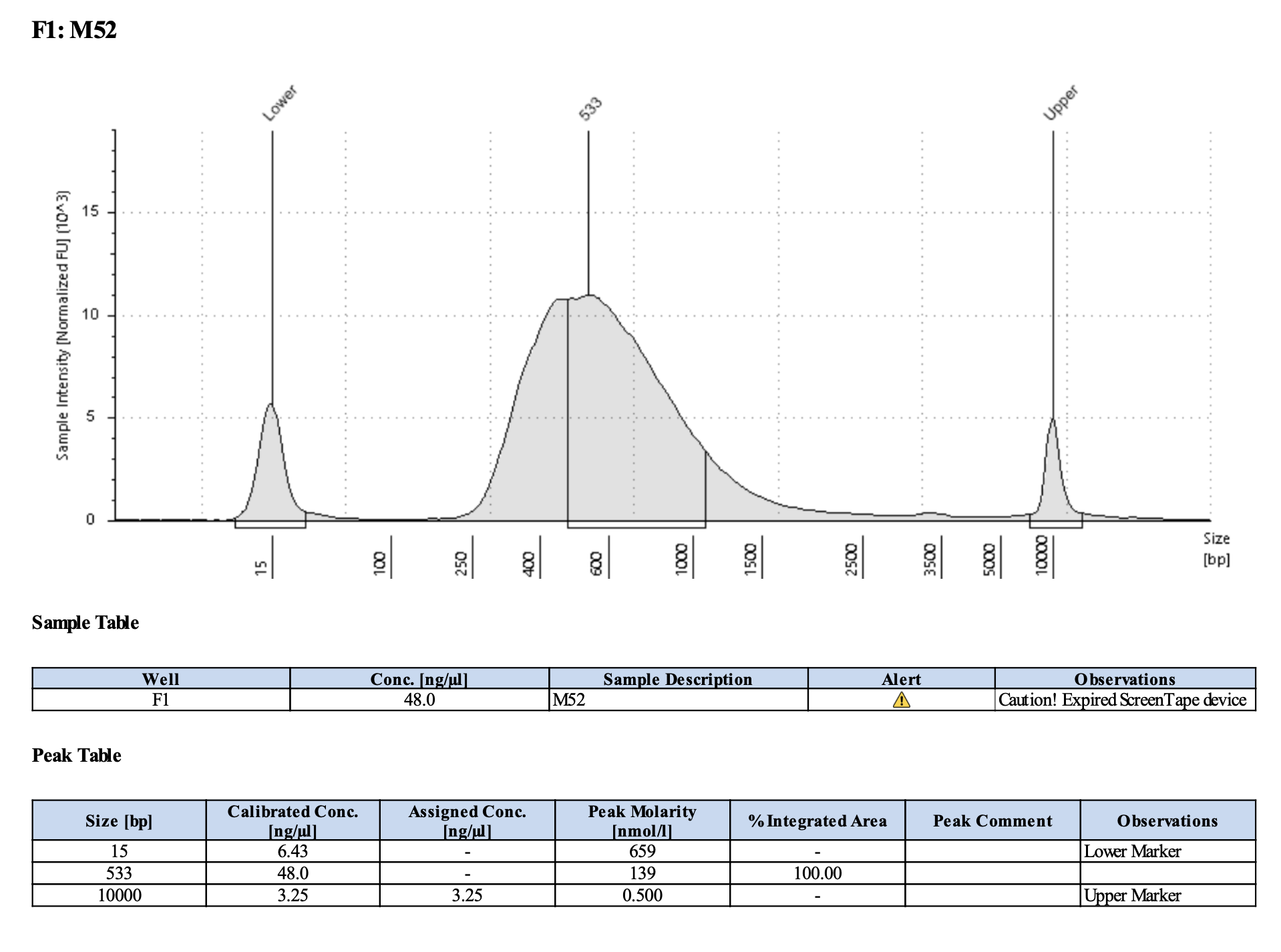
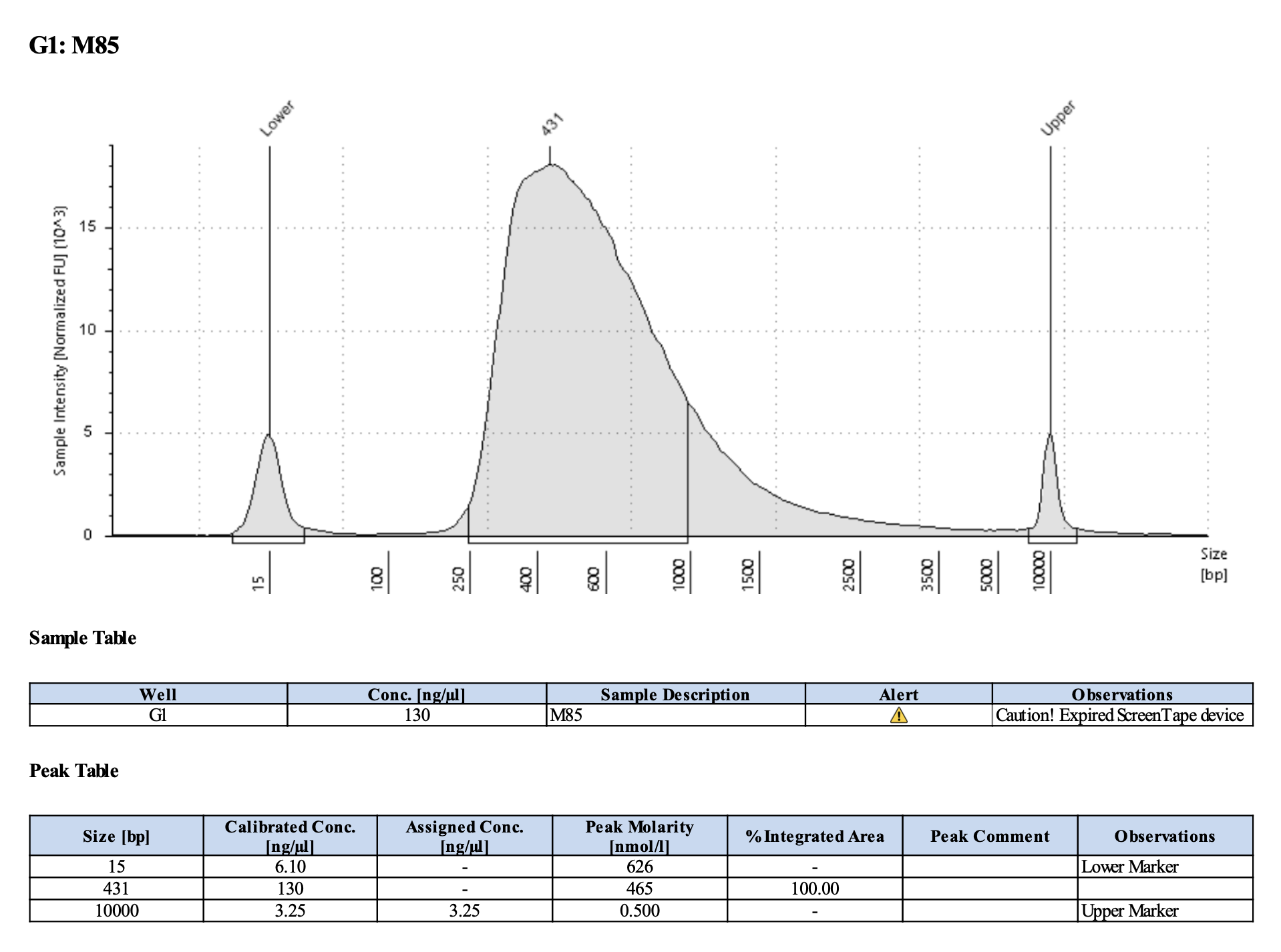
Peaks are similar in height and width to my previous prep and similar to Maggie’s results in terms of the bp distribution. Overall, I am happy with these libraries and they will be sent for sequencing.