Pico Methyl-Seq library prep
Pico Methy-Seq Library Prep test for E5 samples
This post details the info about the WGBS library prep steps for the e5 deep dive samples collected in Moorea in 2020. The github for that project is linked here. I’m using the Zymo Pico Methyl Seq Library Prep for library prep (we have several kits with 25 preps). See Zymo’s protocol here.
Since there are 3 species for this project (Acropora pulchra, Pocillopora tuahiniensis, and Porites evermanni), I’ll be doing a test of one of each species today. I selected 439 (ACR-173), 403 (POC-48), and 421 (POR-82). The kit needs a minimum input of 10 ng DNA or a maximum input of 100 ng DNA. Emma suggested using 20 ng, as that amount has yielded better libraries than 10 ng, so I’ll go with 25 ng. Both Maggie and Emma have done this protocol before with success.
| Number | colony_id | Species | DNA average (ng/uL) | DNA input (uL) | Tris (uL) | Total starting volume | Primer |
|---|---|---|---|---|---|---|---|
| 439 | ACR-173 | Acropora pulchra | 19.9 | 1.3 | 18.7 | 20 | 20 |
| 403 | POC-48 | Pocillopora tuahiniensis | 29.1 | 0.9 | 19.1 | 20 | 21 |
| 421 | POR-82 | Porites evermanni | 3.41 | 7.3 | 12.7 | 20 | 22 |
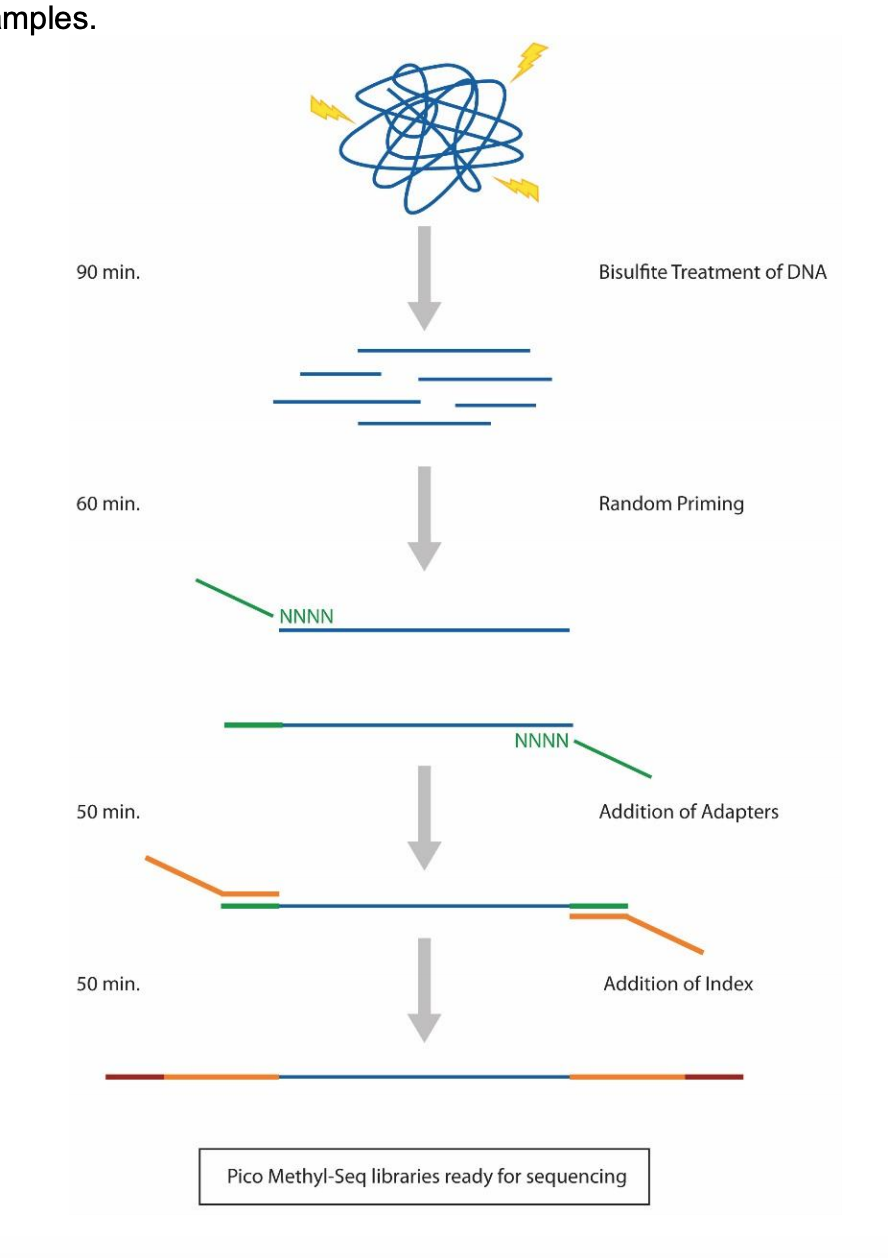
Here’s the Pico Methyl-Seq library prep workflow:
Materials
- Zymo-Seq RiboFree library kit
- PCR tubes
- Thermocycler
- Heating block
- Mini centrifuge
- Vortex
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- 1.5 mL tubes
- Shaker
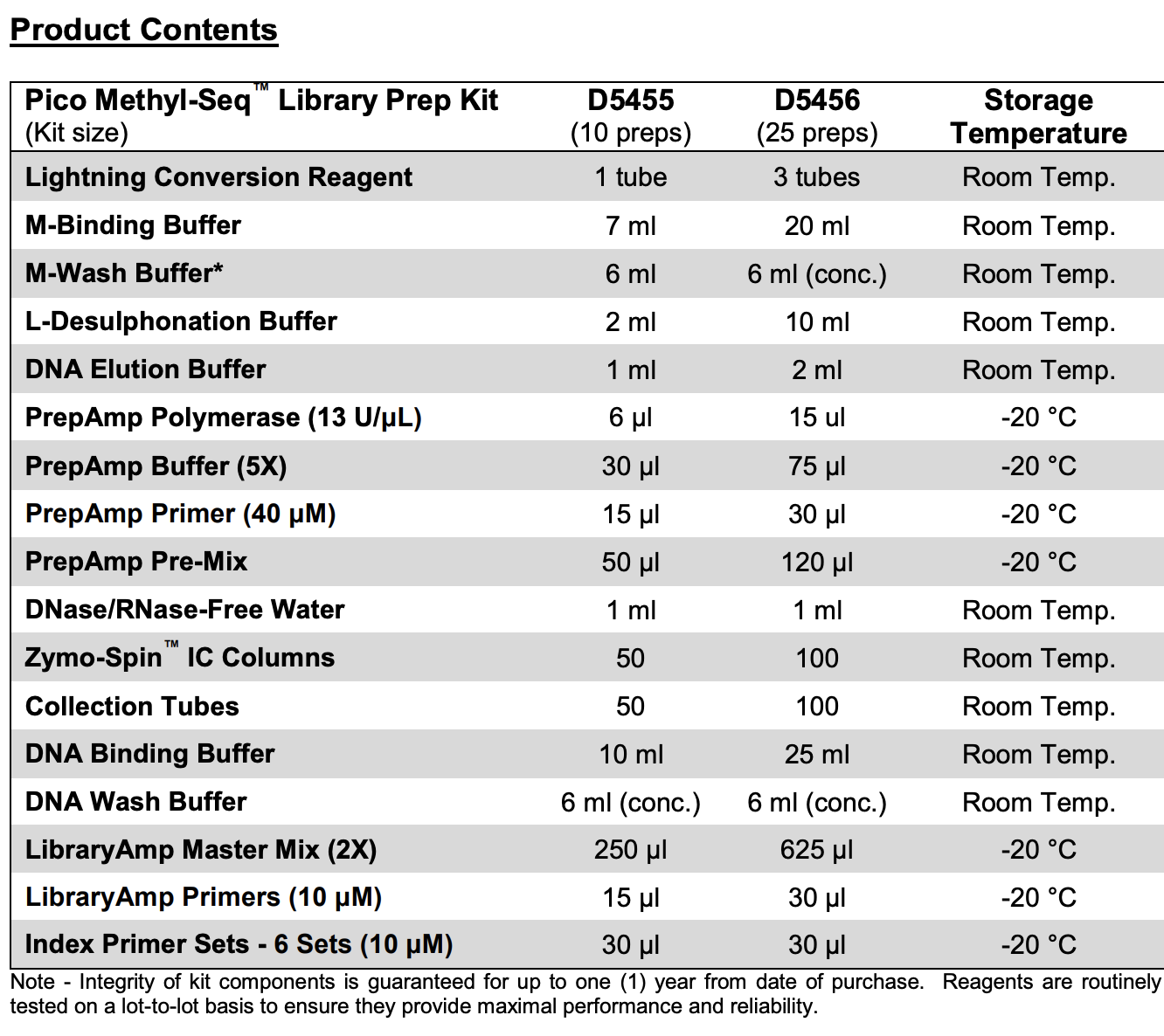
- Kit contents
Buffer preperation
Once buffers are prepared for a kit, they do not need to be prepared again.
- Add 26 mL of 95% EtOH to the M-Wash Buffer concentrate
- Add 26 mL of 95% EtOH to the DNA Wash Buffer concentrate
Best practices
- Preset the thermocycler programs
- Thaw and keep -80°C and -20°C components on ice unless instructed otherwise. Flick to mix and centrifuge before use
- Avoid multiple freeze-thaws, make aliquots if needed
- Calculate master mix volumes before library prepping (see spreadsheet here)
- Allow Kapa beads to equilibrate to room temperature >30 mins before use
- Resuspend magnetic particles immediately before each use by gently inverting until homogenous
Section 1: Bisulfite Conversion of DNA
Thermocycler settings:
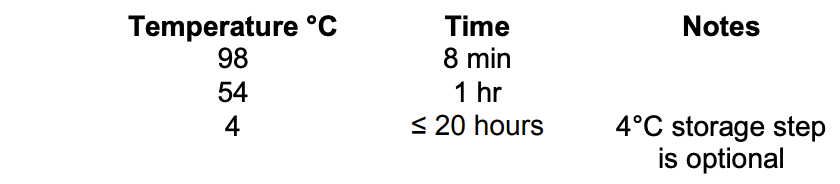
- Thaw samples on ice
- Prepare samples to correct volume in strip tubes
- See table above
- Add 130 uL of Lightning Conversion Reagent to each tube
- Vortex for 10 seconds and centrifuge briefly
- Run thermocycler program (labeled BS CONVERSION PICO under Maggie’s profile)
- Store at 4°C for <20 hours (overnight)
NEXT DAY!
- Label Zymo-spin IC columns, collection tubes and 1.5 mL tubes for each sample
- Add 600 uL of M-Binding Buffer to each column
- Add the entire bisulfite-converted sample to column and invert several times to mix
- Centrifuge at 16,000 g for 30 seconds
- Discard flow through
- Add 100 uL M-Wash Buffer
- Centrifuge at 16,000 g for 30 seconds + discard flow through
- Add 200 uL L-Desulphonation Buffer to column and incubate at room temperature for 15 minutes
- Near end of incubation, heat DNA Elution buffer to 56°C on thermoblock
- After incubation, centrifuge at 10,000 g for 30 seconds + discard flow through
- Add 200 uL of M-Wash Buffer
- Centrifuge at 16,000 g for 30 seconds + discard flow through
- Repeat the wash step above
- Move spin columns into their respective 1.5 mL tubes
- Add 8 uL of warmed DNA Elution buffer to column
- Incubate at room temperature for 5 minutes
- Centrifuge at 16,000 g for 30 seconds to elute bisulfite-converted DNA
- Check if all tubes have same volume (greater elution volume may cause library prep failure)
- Move 1.5 mL tubes to ice
Section 2: Amplification with PrepAmp Primer
Thermocycler settings:
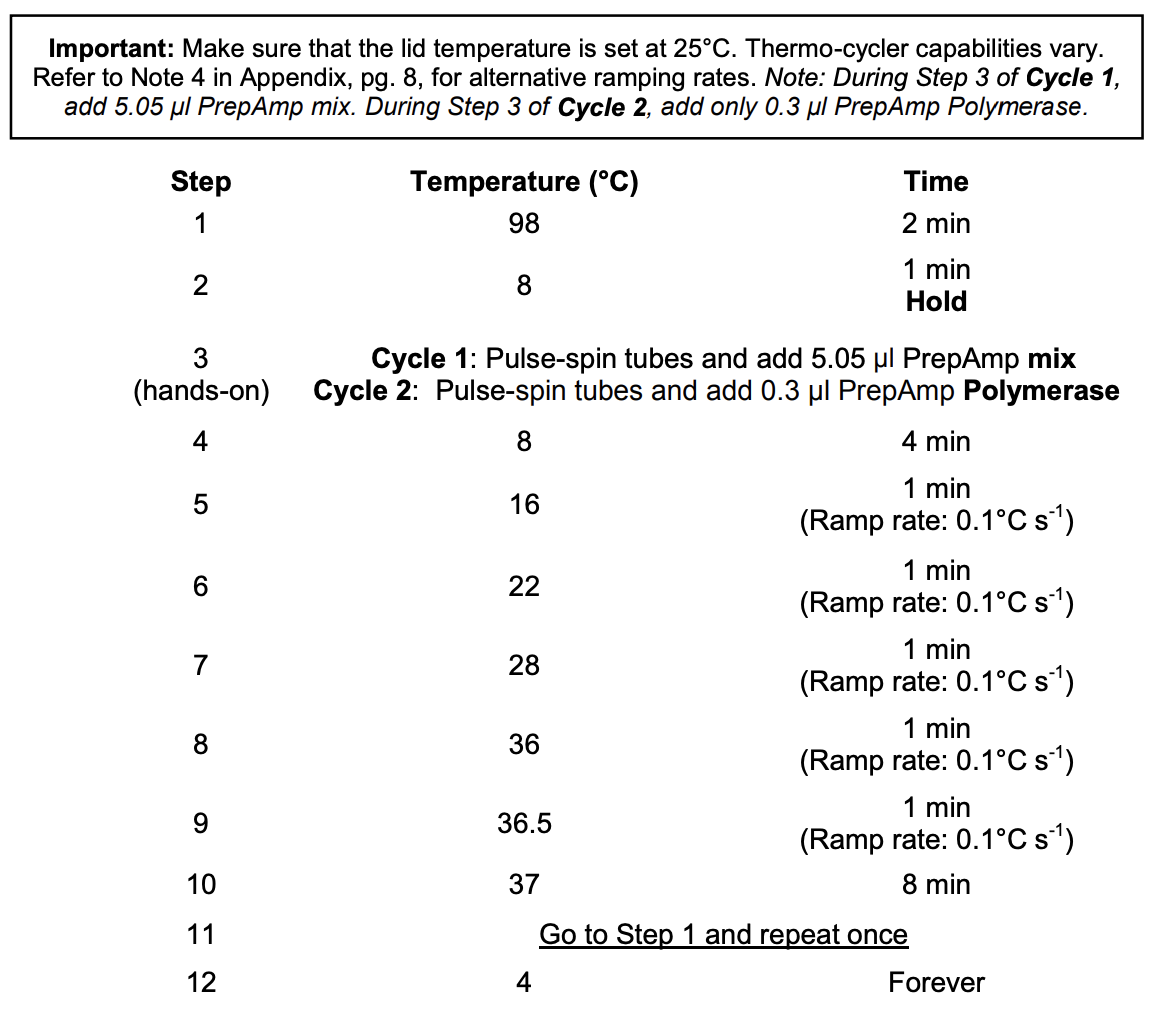
Lid should be set at 25°C
- Thaw the following reagents on ice
- PreAmpBuffer (5x)
- PreAmp Primer (40 uM)
- PreAmp pre-mix
- PreAmp polymerase
- Determine n number: number of samples + % error (~5%)
- Labeling scheme for master mixes
- PMM = A
- PAMM = B
- dPAP = C
- AMM = D
- Create Priming Master Mix (PMM or A) on ice. Vortex and centrifuge briefly
- 2 uL 5x PreAmp Buffer * n =
- 1 uL 40 uM PreAmp Primer * n =
- Create PreAmp Master Mix (PAMM or B) on ice. Vortex and centrifuge briefly
- 1 uL 5x PrepAmp Buffer * n =
- 3.75 uL PrepAmp Pre Mix * n =
- 0.3 uL PrepAmp Polymerase * n =
- Create ‘diluted’ PreAmp Polymerase mix (dPAP or c) on ice. Pipette to mix and centrifuge briefly
- This is to avoid adding less than 0.5 uL during protocol. (original protocol asks you to add 0.3ul to the tubes in the thermocycler, sometimes that small of an amount does not come out of the tip so you can add DNA elution buffer to the enzyme to pipette 0.5ul).
- 0.3 uL PrepAmp Polymerase * n =
- 0.2 uL DNA Elution Buffer * n =
- Add 7 uL of bisulfite converted sample to new PCR tubes
- Add 3 uL of PMM (A) to each tube
- Vortex and centrifuge briefly
- Run the thermocycler program with the lid temperature at 25°C with 2 cycles
- During the Step 2 8°C hold:
- Cycle 1: Add 5.05 ul of PreAmp Master Mix (PAMM or B). Pipette up and down to mix, centrifuge briefly, put back in thermocycler and proceed past hold
- Cycle 2: Add 0.5 uL of diluted PreAmp Polymerase (dPAP or C) to each tube.Pipette up and down to mix, centrifuge briefly, put back in thermocycler and proceed past hold
- During the Step 2 8°C hold:
- Put samples on ice
Section 3: Purification with DNA Clean-up and Concentrator (DCC)
- Label 1.5 mL tubes + IC columns for each sample
- Heat DNA Elution Buffer to 56°C on thermoblock
- Add 108.85 uL of DNA binding buffer (7:1 ratio of DNA binding buffer:sample)
- Move the sample from the PCR tube into the 1.5 mL tube
- Vortex and spin down to mix
- Move sample into a Zymo-Spin IC column
- Centrifuge at 16,000 g for 30 seconds + discard flow through
- Add 200 uL of DNA Wash Buffer to each column
- Centrifuge at 16,000 g for 30 seconds + discard flow through
- Repeat this wash step
- Move spin columns into their respective 1.5 mL tubes
- Add 12 uL of warmed DNA Elution Buffer
- Incubate spin columns at room temperature for 5 minutes
- Centrifuge at 16,000 g for 30 seconds
- Toss spin columns and keep 1.5 mL tubes on ice
Section 4: Library Amplification
Thermocycler settings:
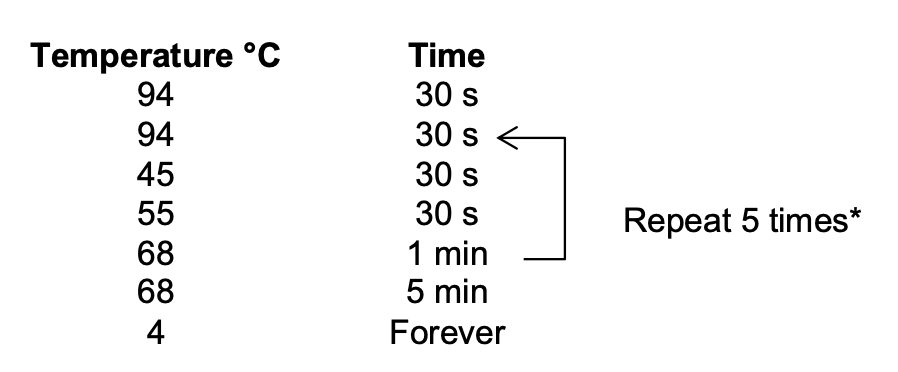
I will do 8 total cycles instead of 6 (based off of Maggie and Emma’s experience).
- Make Amplification Master Mix (AMM or D) on ice. Vortex and centrifuge briefly
- 12.5 uL 2X Library Amp Mix * n =
- 1 uL 10 uM Library Amp Primers * n =
- Add 13.5 of AMM to new strip tubes
- Add 11.5 uL of sample into each strip tube
- Vortex and centrifuge briefly
- Run the thermocycler program
- Put samples on ice
Section 5: Purification with DNA Clean-up and Concentrator (DCC)
- Label 1.5 mL tubes for each sample
- Heat DNA Elution Buffer to 56°C on thermoblock
- Add 175 uL of DNA binding buffer (7:1 ratio of DNA binding buffer:sample)
- Move the sample from the PCR tube into the 1.5 mL tube
- Move sample into a Zymo-Spin IC column
- Centrifuge at 16,000 g for 30 seconds + discard flow through
- Add 200 uL of DNA Wash Buffer to each column
- Centrifuge at 16,000 g for 30 seconds + discard flow through
- Repeat this wash step
- Move spin columns to 1.5 mL tubes
- Add 12.5 uL of warmed DNA Elution Buffer to the column matrix
- Incubate for 5 minutes at room temperature
- Centrifuge at 10,000 g for 30 seconds to elute
Section 6: Amplification with Index Primers
Thermocycler settings:
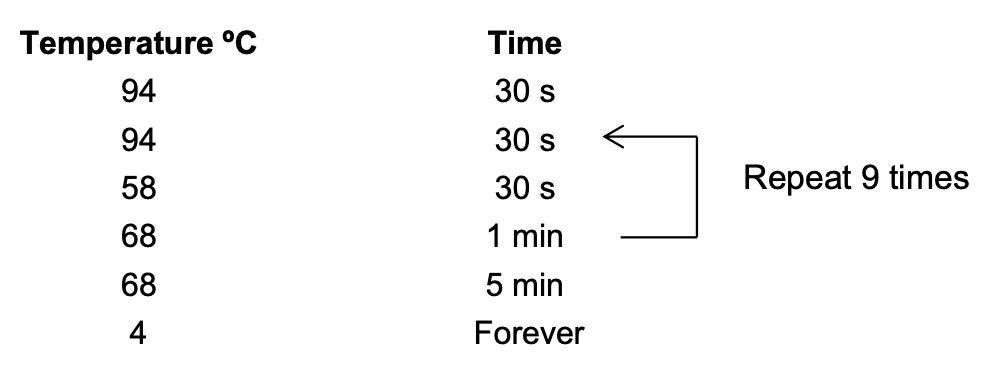
- Move 10.5 uL of sample into new strip tubes
- Add 12.5 uL of 2X Library Amp Master Mix to each sample
- Add 2 uL of combined i7 and i5 primer pair (1 uL of each)
- Run the thermocycler program for 10 amplification cycles
- After program is done, put samples on ice
Section 7: 1x Bead Cleanup
- Take Kapa beads out of fridge >30 minutes before using to allow beads to equilibrate to room temperature
- Shake the bottle to mix the beads (don’t vortex)
- Make fresh 80% ethanol using 100% ethanol
- Add 25 uL of beads to each sample
- Pipette up and down at least 10 times to mix sample and beads (or until homogeous)
- Put strip tubes on the shaker at 200 rpm for 15 minutes
- After 15 minutes, put tubes on magnetic stand
- Wait until liquid is clear (~3-5 minutes)
- Using a p200 set to 45 uL, remove the supernatent without disturbing the beads and discard
- Add 200 uL of 80% ethanol to each sample
- Remove ethanol from tube without disturbing beads and discard liquid
- Repeat the wash step
- Allow beads to air dry for 1-2 minutes
- Remove any residual liquid from each tube
- Resuspend beads in 15 uL of DNA Elution Buffer
- Put tubes back on shaker at 200 rpm for 5 minutes
- After 5 minutes, put tubes back on magnetic stand and wait until liquid is clear (~3-5 minutes)
- Move 15 uL of elute into new strip tubes
THIS IS THE FINAL WGBS LIBRARY. STORE AT -20°C.
QC
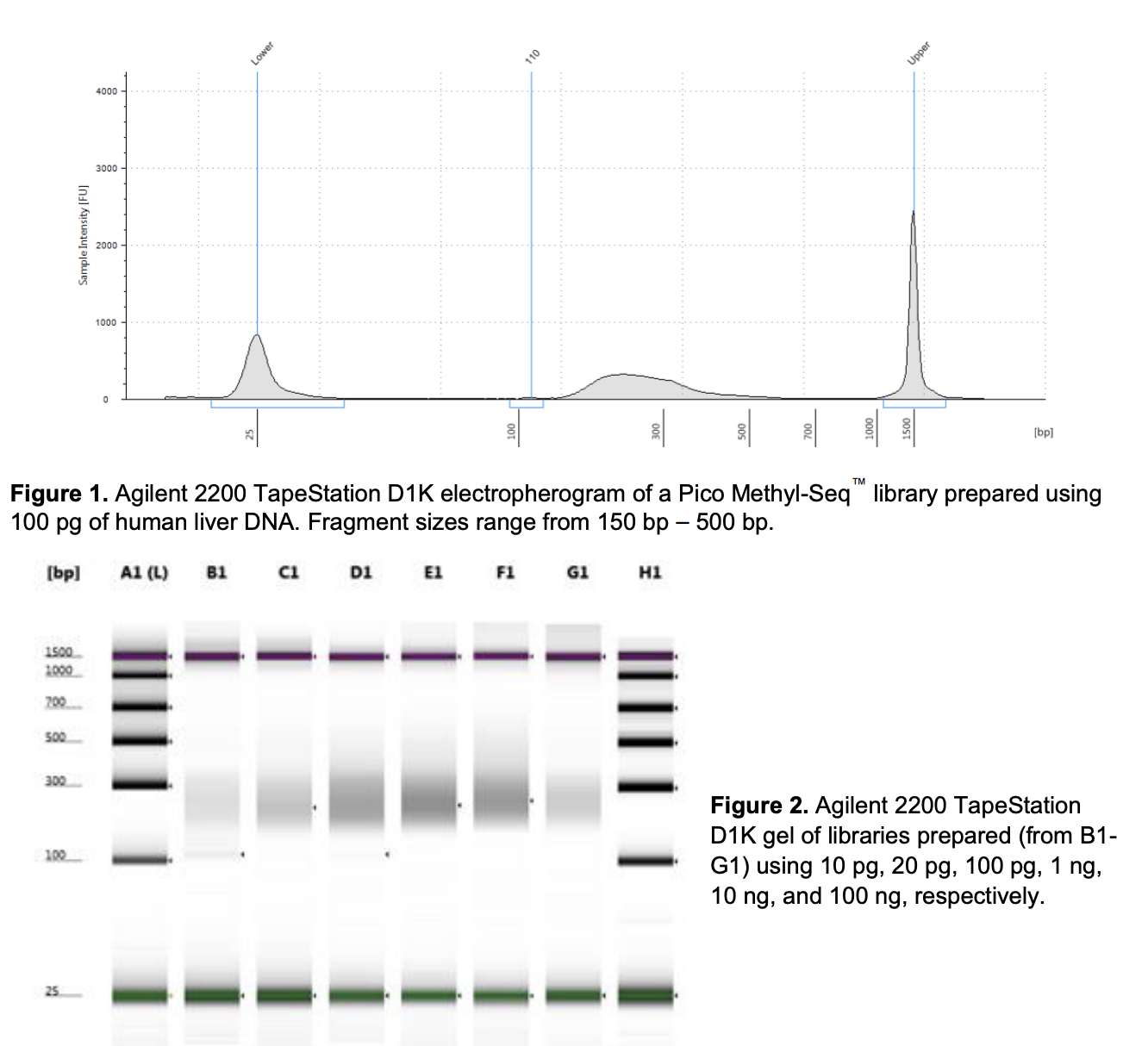
Run DNA Tapestation for visualize libraries. Here’s an example of what the library should look like on a Tapestation: