Pico Methyl-Seq library prep
Pico Methy-Seq Library Prep for E5 samples
This post details the info about the WGBS library prep steps for the e5 deep dive samples collected in Moorea in 2020. The github for that project is linked here. I’m using the Zymo Pico Methyl Seq Library Prep for library prep (we have several kits with 25 preps). See Zymo’s protocol here.
I prepped the 15 deep dive E5 samples in June 2024 (see notebook post) but 6 samples failed to amplify (3 Pocillopora tuahiniensis and 3 Porites evermanni). I re-tried the 6 samples that failed in August 2024 (see notebook post) but these failed as well. I got slight peaks for the POC samples but concentrations were low (<1ng/ul). I re-tried again with these samples in late August 2024 (see notebook post) but these failed once again, depsite increasing the number of cycles for POC samples and decreasing the DNA input amount for POR samples.
Going to retry one more time with these samples. For POC samples, I will increase the DNA input amount slightly (35-50ng) and increase the number of cycles to 15. For the POR samples, I will use the cleaned samples (see notebook post) and keep the number of cycles the same (10). I’m using extractions done by myself (9/25 date) and Kristina (2021 dates). I am also running one of Zoe’s samples from her LCM experiment.
| Number | Species | Timepoint | Collection Date | Site | colony_id | Extraction Date | Extraction notebook post | DNA (ng/uL) | Volume eluted (uL) | Total DNA (ng) | Starting volume (uL) | Volume for DNA (uL) | Tris (uL) | DNA input amount (ng) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 385 | Pocillopora tuahiniensis | TP2 | 20200305 | Site1 | POC-47 | 20211012 | https://kterpis.github.io/Putnam_Lab_Notebook/20211012-RNA-DNA-extractions-from-E5-project/ | 25.5 | 90 | 2295.00 | 20 | 2.0 | 18.0 | 51 |
| 393 | Pocillopora tuahiniensis | TP2 | 20200305 | Site1 | POC-50 | 20210903 | https://github.com/Kterpis/Putnam_Lab_Notebook/blob/master/_posts/2021-09-03-20210903-RNA-DNA-extractions-from-E5-project.md | 29.4 | 90 | 2646.00 | 20 | 2.0 | 18.0 | 58.8 |
| 401 | Pocillopora tuahiniensis | TP2 | 20200305 | Site1 | POC-53 | 20211118 | https://kterpis.github.io/Putnam_Lab_Notebook/20211118-RNA-DNA-extractions-from-E5-project/ | 30.8 | 90 | 2772.00 | 20 | 2.0 | 18.0 | 61.6 |
| 421 | Porites evermanni | TP2 | 20200305 | Site1 | POR-82 | 20211008 | https://kterpis.github.io/Putnam_Lab_Notebook/20211008-RNA-DNA-extractions-from-E5-project/ | 0.580 | 80 | 46.40 | 20 | 20 | 0 | 11.6 |
| 487 | Porites evermanni | TP2 | 20200305 | Site1 | POR-71 | 20211122 | https://github.com/Kterpis/Putnam_Lab_Notebook/blob/master/_posts/2021-11-22-20211122-RNA-DNA-extractions-from-E5-project.md | 0.954 | 80 | 76.32 | 20 | 20 | 0 | 19.08 |
| 489 | Porites evermanni | TP2 | 20200305 | Site1 | POR-79 | 20211129 | https://github.com/Kterpis/Putnam_Lab_Notebook/blob/master/_posts/2021-11-29-20211129-RNA-DNA-extractions-from-E5-project.md | 0.804 | 80 | 64.32 | 20 | 20 | 0 | 16.08 |
| 421 | Porites evermanni | TP2 | 20200305 | Site1 | POR-82 | 20240925 | https://github.com/JillAshey/JillAshey_Putnam_Lab_Notebook/blob/master/_posts/2024-09-25-MiniprepPlus-DNA-extractions-E5.md | 0.116 | 75 | 8.70 | 20 | 20 | 0 | 2.32 |
| 487 | Porites evermanni | TP2 | 20200305 | Site1 | POR-71 | 20240925 | https://github.com/JillAshey/JillAshey_Putnam_Lab_Notebook/blob/master/_posts/2024-09-25-MiniprepPlus-DNA-extractions-E5.md | 0.240 | 75 | 18.00 | 20 | 20 | 0 | 4.8 |
| 489 | Porites evermanni | TP2 | 20200305 | Site1 | POR-79 | 20240925 | https://github.com/JillAshey/JillAshey_Putnam_Lab_Notebook/blob/master/_posts/2024-09-25-MiniprepPlus-DNA-extractions-E5.md | 0.141 | 75 | 10.58 | 20 | 20 | 0 | 2.82 |
| 13 | Pocillopora acuta ZD | NA | NA | NA | NA | 20240908 | https://zdellaert.github.io/ZD_Putnam_Lab_Notebook/LCM-Exp-Test-Extraction/ | See ZD notebook | See ZD notebook | See ZD notebook | 20 | 5 | 15 | See ZD notebook |
Here’s the Pico Methyl-Seq library prep workflow:
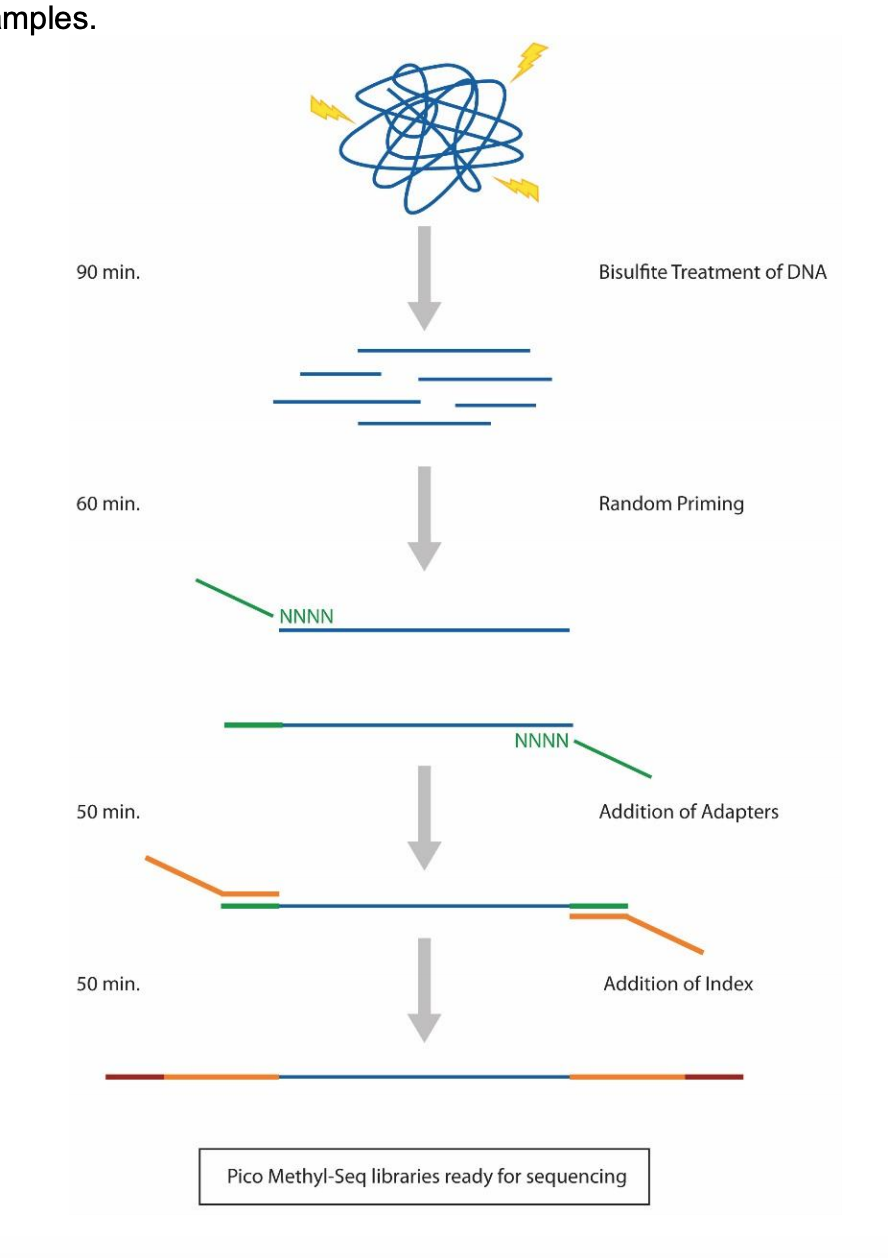
Protocol
I followed this protocol with a few modifications:
- Section 1 - incubation with warmed DNA elubtion buffer shortened from 5 to 3 minutes. Emma reported that she typically got less out of the column if she did the full 5 minutes.
- Section 3 - Incubation with warmed DNA elubtion buffer shortened from 5 to 2.5 minutes.
- Section 4 - number of cycles for POC was 15; number of cycles for POR was 10.
- Section 5 - incubation with warmed DNA elubtion buffer shortened from 5 to 2.5 minutes.
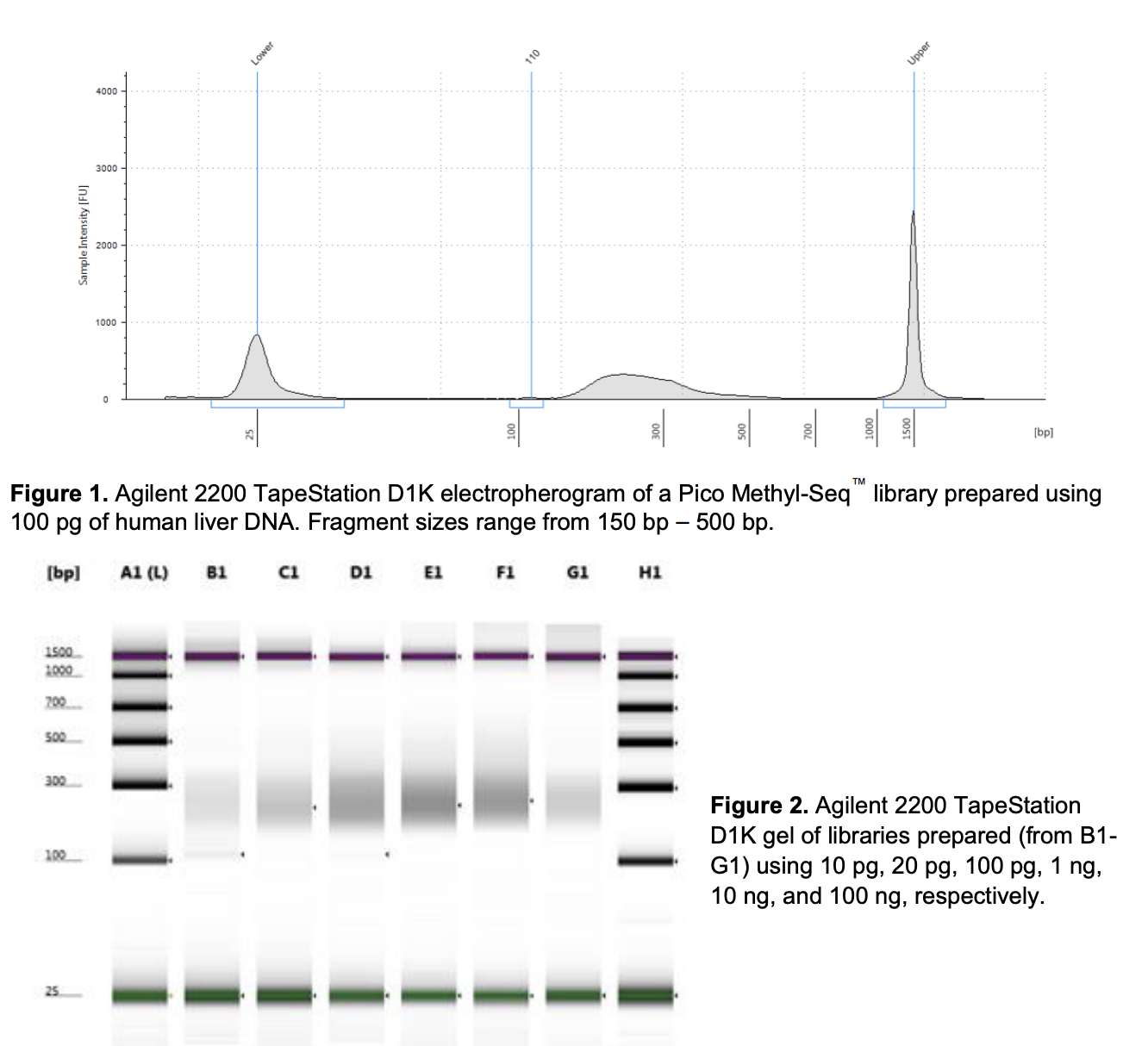
QC
Run DNA Tapestation for visualize libraries. Here’s an example of what the library should look like on a Tapestation:
Here’s what my libraries looked like on 10/15/24. See full results here.
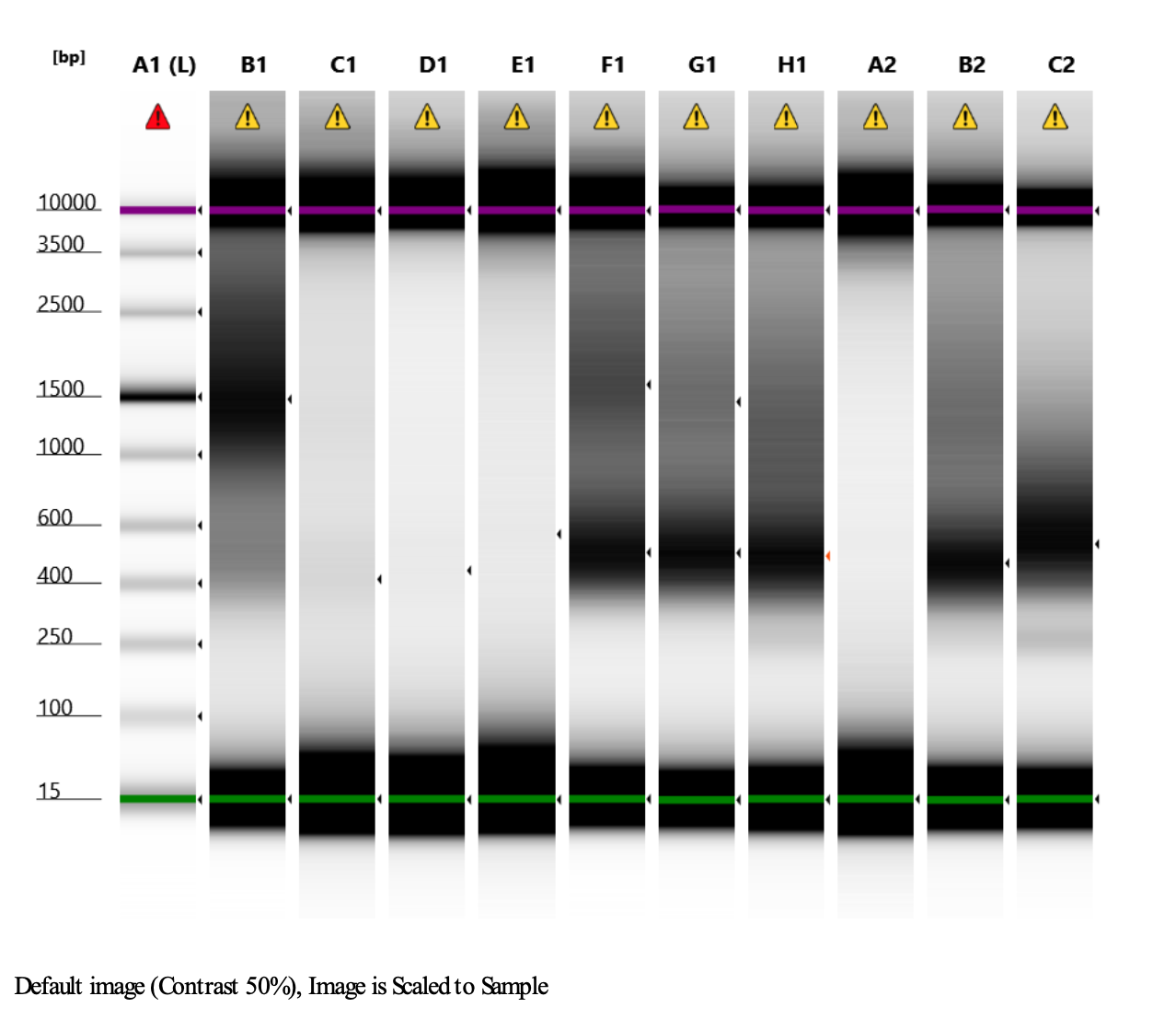
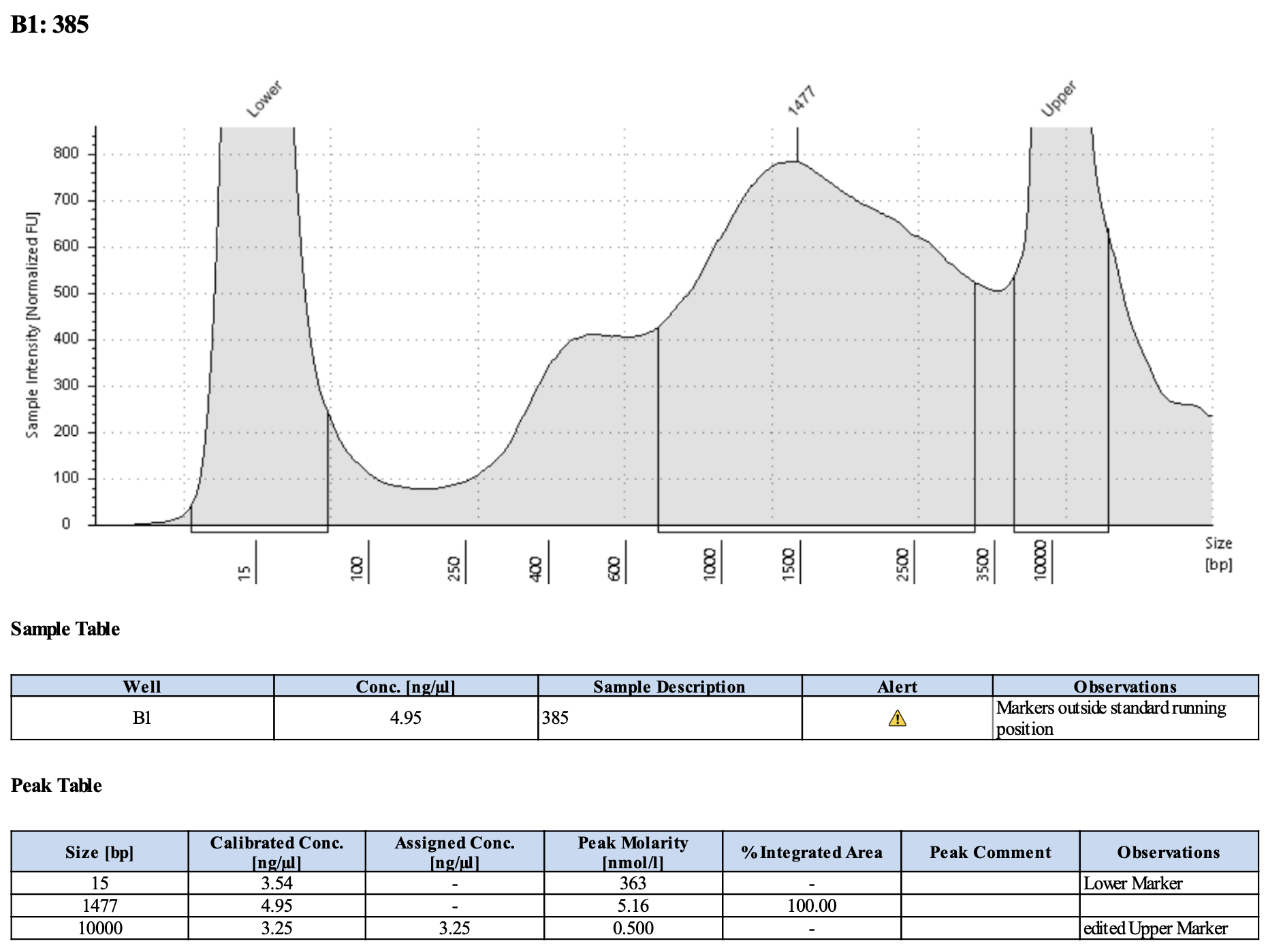
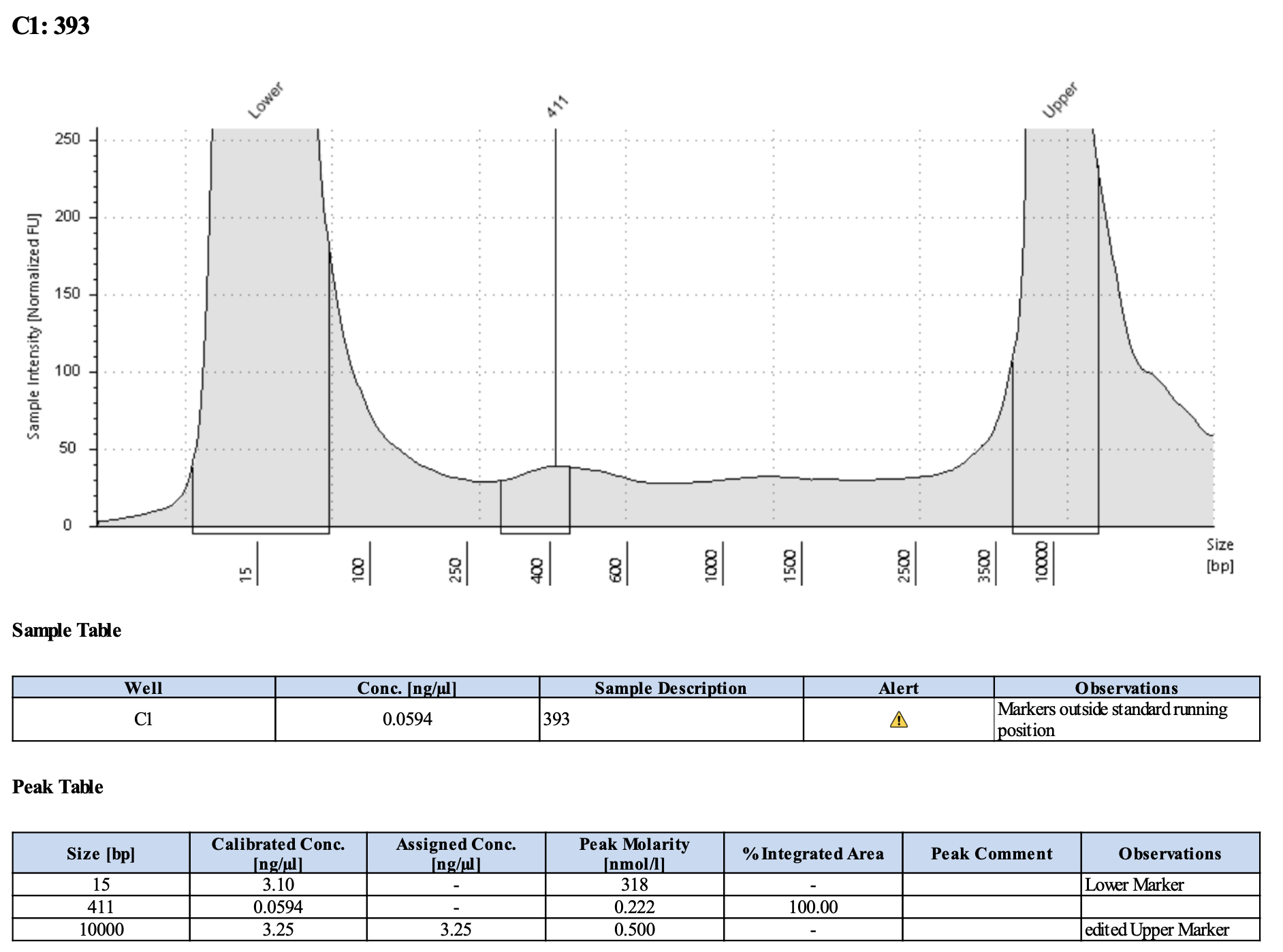
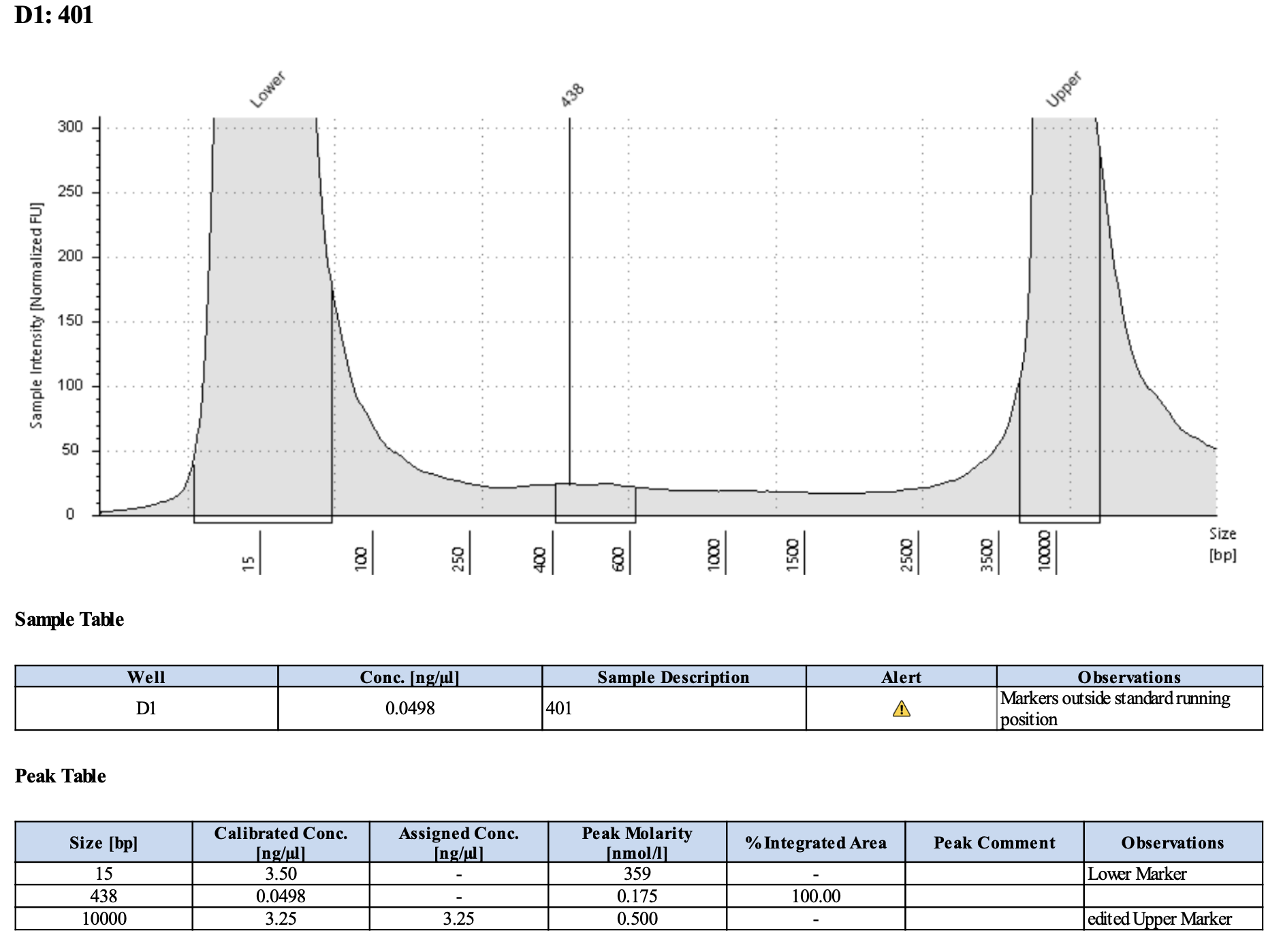







Mine are really all over the place. Zoe’s sample worked!!