MiniPrep Plus DNA/RNA extractions
Extractions for Mcap developmental time series Hawaii 2023
This protocol is based on Maggie’s coral egg/larvae extraction protocol and Kevin’s coral larvae extraction protocol. This post details the info about the extraction steps for the M. capitata developmental time series experiment in Hawaii 2023. The github for that project is linked here.
Samples
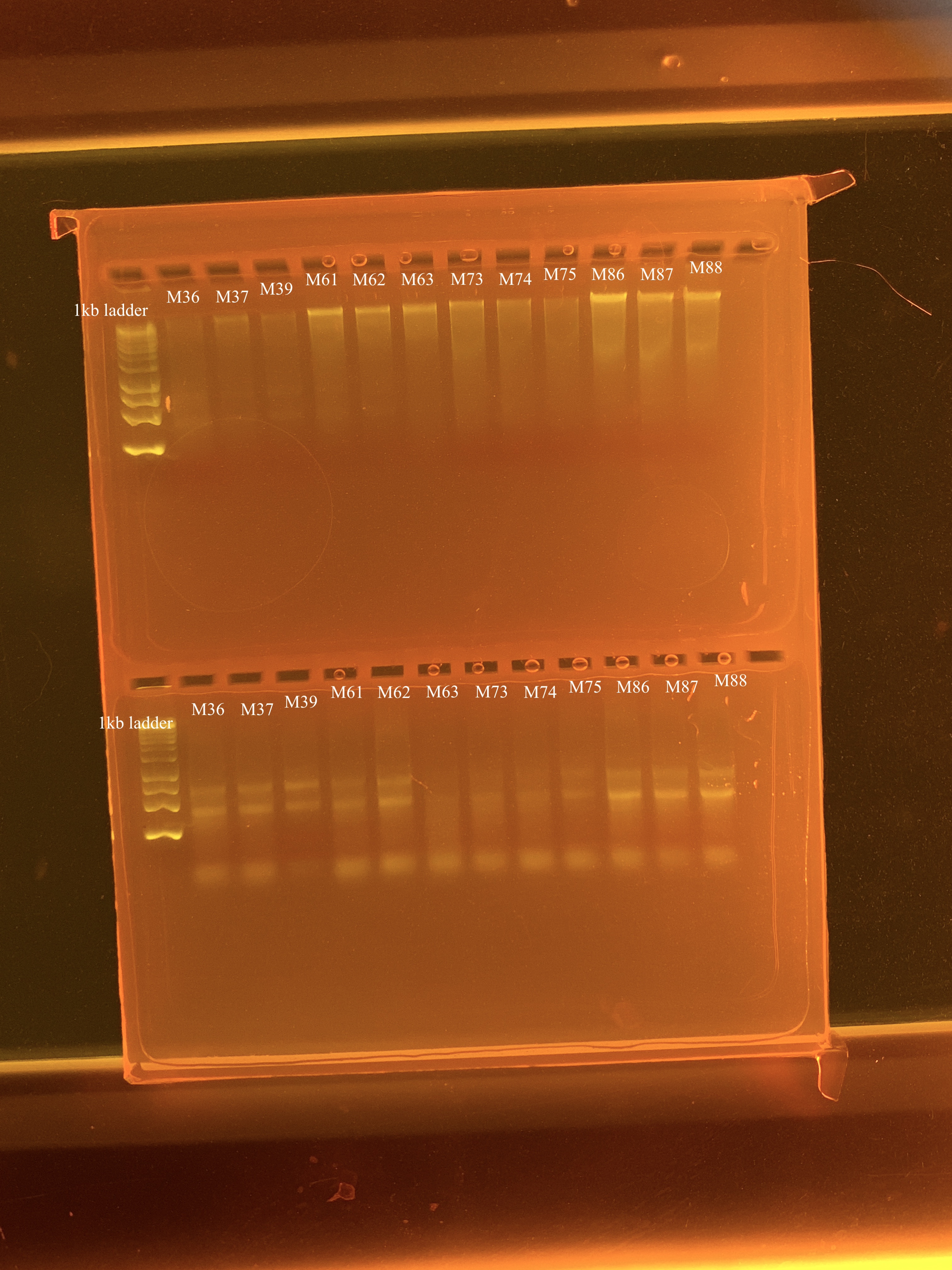
The samples run today were a mix of new and previously extracted samples. M37, M38, M39, M73, M74, M75, M86, M87, and M88 were all new. I did not add beads to these samples for beating step and I used 300uL as my input volume instead of 500uL. See the bottom of this post for reasoning behind those choices.
M61 was originally extracted on 1/5/24, and M63 and M63 were originally extracted on 2/8/24. These samples were originally extracted using the bead beating method and taking 500 uL as input. In this round of (re)extractions, I took 300 uL of input from these samples. These samples had already been bead beaten from previous extractions.
Forgot to take sample picture :’(
Materials
- Zymo Quick-DNA/RNA Miniprep Plus Kit HERE Protocol Booklet
- Tris-Ethylenediaminetetraacetic acid (EDTA) 1X buffer for DNA elution
- Heating block capable of heating to 70ºC
- Centrifuge and rotor capable of spinning at 15,000 rcf
- Plastics
- 5 1.5 mL microcentrifuge tubes per sample
- 2 PCR tubes per sample
- 2 Qubit tubes per sample
Protocol
Protocol was followed according to this post. A few differences:
- I added between 200-500 uL of DNA/RNA shield to the new samples that had <1mL of shield in them after thawing them so that the volume in the tubes was 1 mL of shield. I aliquoted out 300 uL for extractions and saved the remaining fraction in case we need to re-extract.
- I did not add beads when mixing the samples after they thawed for the newly extracted samples. Samples that were being re-extracted already had beads.
- In the RNA extraction, I added another wash step with 400 uL of wash buffer after the DNase incubation (i.e., after the incubation, I did 400 uL of prep buffer, 700 uL of wash buffer, 400 uL of wash buffer, and another 400 uL of wash buffer).
- I eluted in 80 uL of either Tris (DNA) or DNA/RNA free water (RNA) instead of 100 uL.
QC
Qubit results
| Tube ID | Qubit DNA1 | Qubit DNA2 | Qubit DNA Average | Qubit RNA1 | Qubit RNA2 | Qubit RNA Average |
|---|---|---|---|---|---|---|
| M36 | 2.38 | 2.32 | 37 | 36.6 | 36.8 | |
| M37 | 2 | NA | 2 | 34.8 | 35 | 34.9 |
| M39 | NA | NA | NA | 21.6 | 21 | 21.3 |
| M62 | 6.56 | 6.52 | 6.54 | 45 | 45 | 45 |
| M63 | 5 | 4.92 | 4.96 | 30 | 29.6 | 29.8 |
| M61 | 10.6 | 10.5 | 10.55 | 30 | 29.6 | 29.8 |
| M73 | 11 | 10.9 | 10.95 | 30.8 | 30.6 | 30.7 |
| M74 | 6.48 | 6.46 | 6.47 | 32.4 | 32.4 | 32.4 |
| M75 | 6.36 | 6.3 | 6.33 | 27.6 | 27.2 | 27.4 |
| M86 | 44.8 | 44.4 | 44.6 | 75.8 | 76 | 75.9 |
| M87 | 28.6 | 28.4 | 28.5 | 71.8 | 71.4 | 71.6 |
| M88 | 38.8 | 38.2 | 38.5 | 97.2 | 96.8 | 97 |