MiniPrep Plus DNA/RNA extractions
Extractions for Mcap larvae C/D Hawaii 2023 experiment
This protocol is based on Maggie’s coral egg/larvae extraction protocol and Kevin’s coral larvae extraction protocol. This post details the info about the extraction steps for the test samples that were collected in HI 2023 for A. Huffmyer’s experiment on symbiont identity on Montipora capitata coral larval thermal performance. The github for that project is linked here.
Samples
The samples run today were re-extractions for samples R56, R59, R60, R64, R68, R69, R74, R77, R79, R81, R88, R94, R96, R98, R99, R104, R105, R106, and R107. In the original extraction, these samples had 50 larvae in them and were preserved with 700 uL of DNA/RNA Shield. 300 uL of shield was added to them and 500 uL was subsampled for potential re-extraction. The 500 uL subsampled fraction was used today. This photo is post-bead beating:
Materials
- Zymo Quick-DNA/RNA Miniprep Plus Kit HERE Protocol Booklet
- Tris-Ethylenediaminetetraacetic acid (EDTA) 1X buffer for DNA elution
- Heating block capable of heating to 70ºC
- Centrifuge and rotor capable of spinning at 15,000 rcf
- Plastics
- 3 1.5 mL microcentrifuge tubes per sample
- 2 PCR tubes per sample
- 2 Qubit tubes per sample
- 1 5 mL tube per sample
Protocol
Protocol was followed according to this post. A few differences:
- I used the 500 uL subsample fraction for the re-extraction
- In the RNA extraction, I added another wash step with 400 uL of wash buffer after the DNase incubation (i.e., after the incubation, I did 400 uL of prep buffer, 700 uL of wash buffer, 400 uL of wash buffer, and another 400 uL of wash buffer).
- I eluted in 80 uL of either Tris (DNA) or DNA/RNA free water (RNA) instead of 100 uL.
- I used the High Sensitivity RNA Qubit kit for QC instead of the Broad Range kit.
QC
QC for these samples was done on 1/2/24.
Qubit & Nanodrop
| Sample ID | Qubit DNA1 | Qubit DNA2 | Qubit DNA average | Nanodrop DNA concentration | Nanodrop DNA 260/280 | Nanodrop DNA 260/230 | Qubit RNA1 | Qubit RNA2 | Qubit RNA average | Nanodrop RNA concentration | Nanodrop RNA 260/280 | Nanodrop RNA 260/230 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R104 | 2.52 | 2.66 | 2.59 | 5.3 | 1.81 | 0.44 | 13.1 | 13.3 | 13.2 | 10.9 | 2.34 | 1.3 |
| R56 | NA | NA | NA | 6 | 1.63 | 1.3 | 17.7 | 17.4 | 17.55 | 9.2 | 2.62 | 0.26 |
| R81 | NA | NA | NA | 2.8 | 1.86 | 0.56 | 24.2 | 23.6 | 23.9 | 15.2 | 2.14 | 1.13 |
| R107 | 3.68 | 3.7 | 3.69 | 5.2 | 1.67 | 0.89 | 17 | 16.7 | 16.85 | 11.7 | 2.39 | 1.39 |
| R59 | 6.62 | 6.7 | 6.66 | 10.7 | 1.82 | 1.62 | 17.7 | 17.8 | 17.75 | 10.7 | 2.43 | 0.9 |
| R69 | NA | NA | NA | 3 | 1.55 | 1.08 | 20.2 | 19.9 | 20.05 | 15.2 | 2.47 | 0.05 |
| R79 | 6.64 | 6.78 | 6.71 | 15.1 | 1.65 | 1.35 | 40.6 | 40.2 | 40.4 | 27.3 | 2.19 | 1.46 |
| R94 | 4.62 | 4.5 | 4.56 | 8.9 | 1.75 | 1.92 | 31.4 | 30.8 | 31.1 | 23 | 2.13 | 1.23 |
| R99 | NA | NA | NA | 4.7 | 1.85 | 0.89 | 21.6 | 21.6 | 21.6 | 14.1 | 2.19 | 1.29 |
| R106 | 2.82 | 2.9 | 2.86 | 10.8 | 1.74 | 1.41 | 24.2 | 24.6 | 24.4 | 15.8 | 2.38 | 1.41 |
| R60 | NA | NA | NA | 6.3 | 1.82 | 1.62 | 23.4 | 22.8 | 23.1 | 12.2 | 2.36 | 1.1 |
| R64 | 8.86 | 8.52 | 8.69 | 9 | 1.88 | 19.25 | 28.2 | 27 | 27.6 | 16.7 | 2.4 | 1.31 |
| R68 | 4.42 | 4.26 | 4.34 | 9.7 | 1.86 | 0.03 | 32.2 | 31.6 | 31.9 | 22.6 | 2.13 | 1.11 |
| R74 | 5.22 | 5.38 | 5.3 | 9.3 | 1.75 | 0.98 | 23.6 | 23.4 | 23.5 | 20.8 | 2.1 | 1.01 |
| R88 | 4.36 | 4.36 | 4.36 | 7.8 | 2.11 | 0.06 | 24.4 | 24 | 24.2 | 23.4 | 2.09 | 0.79 |
| R96 | 2.78 | 2.68 | 2.73 | 5.6 | 1.62 | 0.43 | 21.2 | 21.2 | 21.2 | 13.7 | 2.3 | 1.24 |
| R98 | NA | NA | NA | 5.2 | 2.13 | 0.02 | 22.8 | 22.4 | 22.6 | 15.4 | 2.3 | 1.41 |
| R77 | 3.18 | 2.7 | 2.94 | 6.9 | 1.7 | 0.57 | 21.2 | 20.6 | 20.9 | 15.7 | 2.17 | 0.72 |
| R105 | 7.54 | 7.4 | 7.47 | 17.3 | 1.71 | 1.55 | 24.6 | 24.4 | 24.5 | 17.1 | 2.33 | 1.26 |
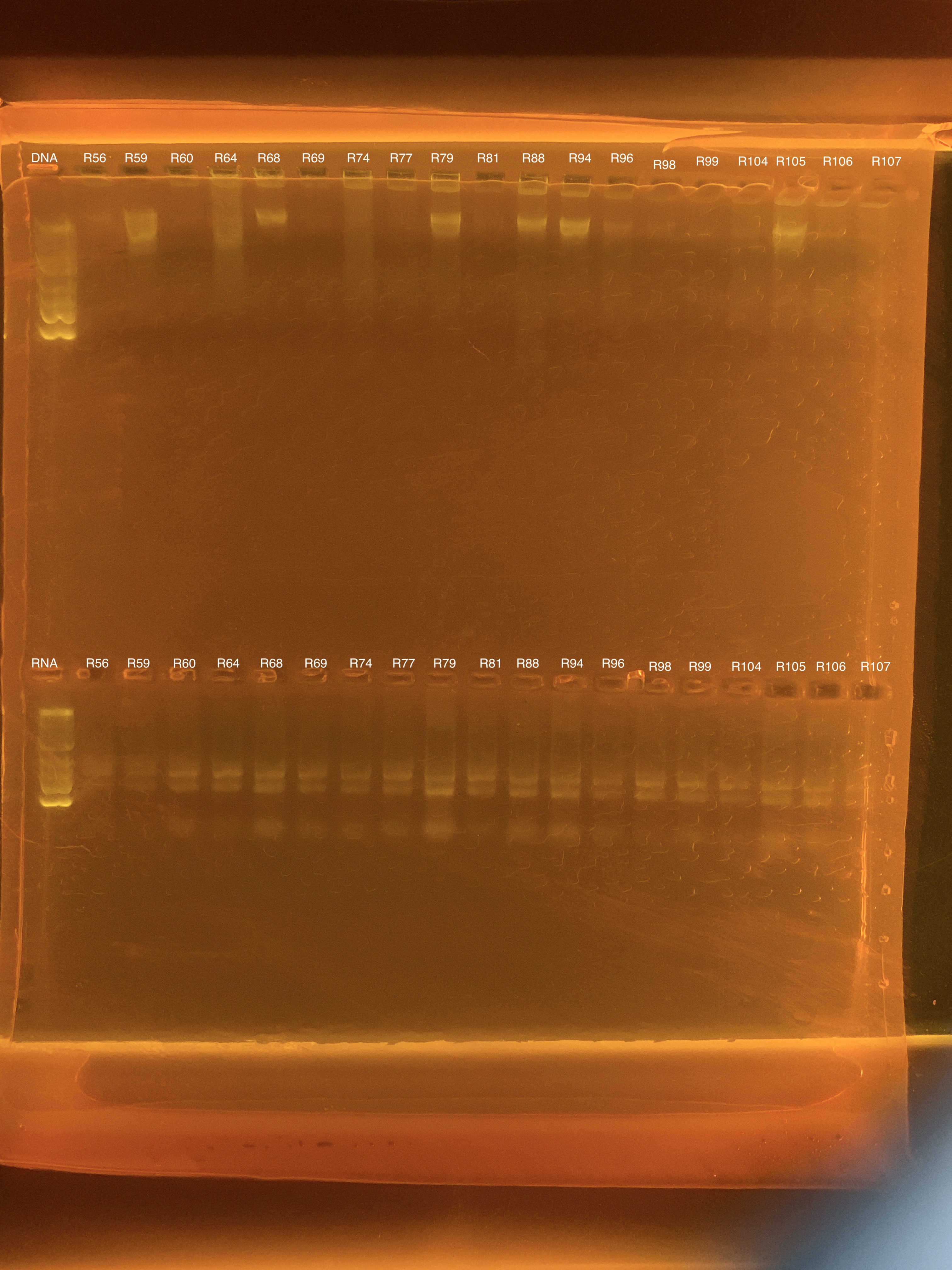
Gel
Several samples had no detectable DNA and there are no bands in the gel for many of the DNA samples. RNA all had good concentrations and bands can be seen, hooray!