MiniPrep Plus DNA/RNA extractions
Extractions for e5 time series
This protocol is based on the Putnam lab Zymo Miniprep protocol (see example from Zoe here). This post details the info about the extraction steps for 12 samples from the e5 timeseries (Acropora, Pocillopora, Porites spp.) in Moorea 2020. The e5 github is linked here.
Samples
The samples run today were re-extractions from the e5 deep dive samples. We had already sequenced these samples but we are going to sequence them again for the timeseries molecular analysis so we decided to re-extract so there were no batch effects. Since the samples had already been through one extraction, there was <1mL of DNA/RNA shield in the tube.
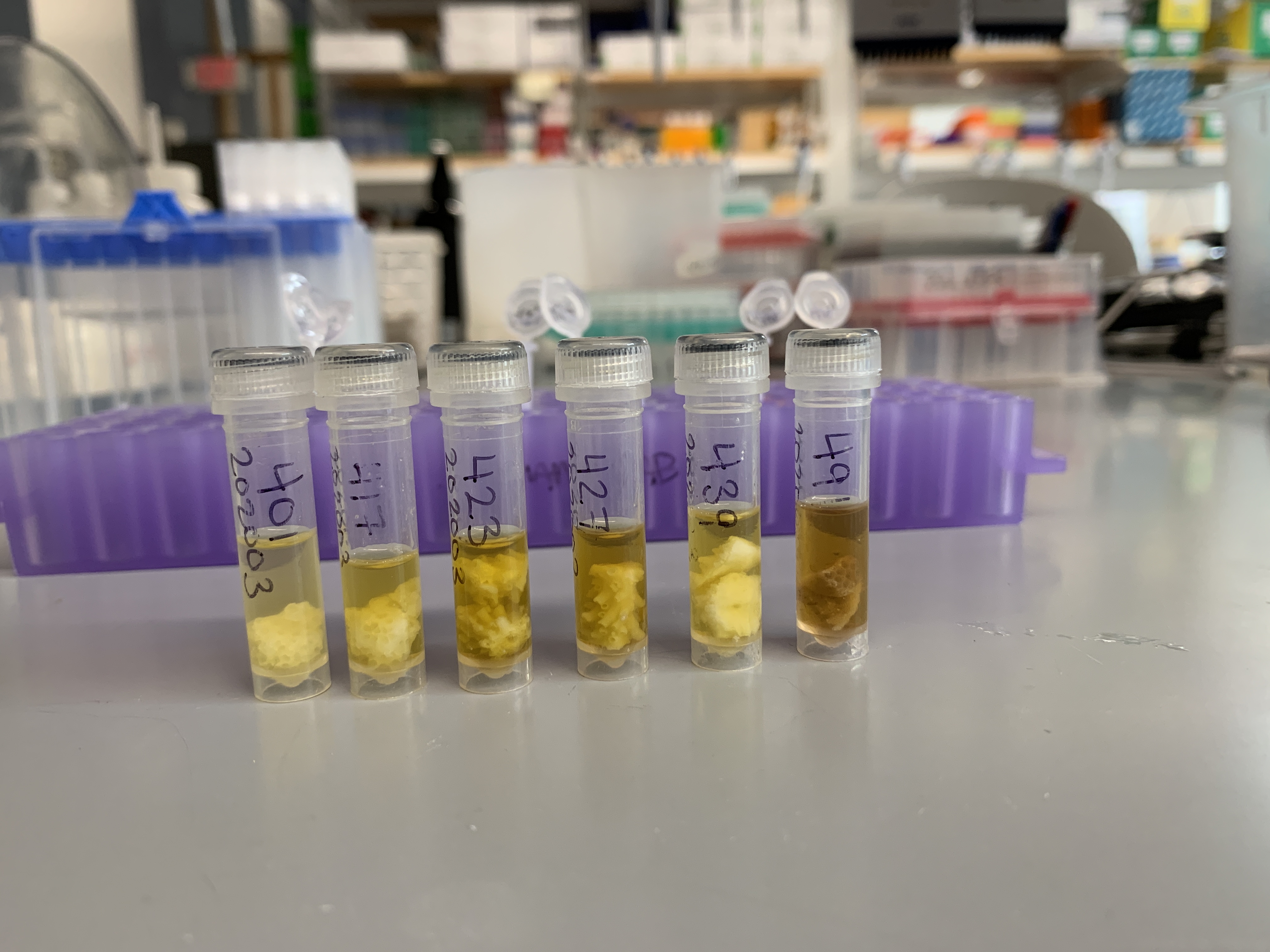
- 401
- 417
- 423
- 427
- 439
- 491
This photo is before I removed shield from the sample tubes:
Materials
- Zymo Quick-DNA/RNA Miniprep Plus Kit HERE Protocol Booklet
- Tris-Ethylenediaminetetraacetic acid (EDTA) 1X buffer for DNA elution
- Heating block capable of heating to 70ºC
- Centrifuge and rotor capable of spinning at 15,000 rcf
- Plastics
- 5 1.5 mL microcentrifuge tubes per sample
- 2 PCR tubes per sample
- 2 Qubit tubes per sample
Protocol
- 300 uL of sample was moved from the original shield tube into a new 1.5mL tube for the prot-k digestion step.
-
For 491, 100 uL of shield + 200 uL of new shield was moved into a new tube. Kristina did this with some of her extractions and it worked well for Porites.
- Samples were snap frozen (in 2020) and clipped into a tube with 1 mL of DNA/RNA shield and 0.5mm glass beads.
- Samples were bead beat for 1.5 minutes on max speed.
This protocol was followed exactly and eluted in 100 uL of either Tris (DNA) or DNA/RNA free water (RNA).
QC
Qubit results
For Qubit, I used the DNA HS kit and the RNA HS kit.
| Shield Tube | Colony ID | Timepoint | Volume | DNA1_ng_uL | DNA2_ng_uL | RNA1_ng_uL | RNA2_ng_uL | DNA_average | RNA_average | DNA_ug | RNA_ug | DNA_ng | RNA_ng |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 401 | POC-53 | TP2 | 90 | 29 | 28.8 | 19.9 | 19.8 | 28.9 | 19.85 | 2.601 | 1.7865 | 2601 | 1786.5 |
| 417 | POC-57 | TP2 | 90 | 77 | 76.2 | 39.8 | 39.4 | 76.6 | 39.6 | 6.894 | 3.564 | 6894 | 3564 |
| 423 | ACR-150 | TP2 | 90 | 36.8 | 36.6 | 14.6 | 14.7 | 36.7 | 14.65 | 3.303 | 1.3185 | 3303 | 1318.5 |
| 427 | ACR-145 | TP2 | 90 | 19.4 | 19.3 | 14 | 13.8 | 19.35 | 13.9 | 1.7415 | 1.251 | 1741.5 | 1251 |
| 439 | ACR-173 | TP2 | 90 | 36.4 | 36.2 | 8.84 | 8.72 | 36.3 | 8.78 | 3.267 | 0.7902 | 3267 | 790.2 |
| 491 | POR-73 | TP2 | 90 | 1.48 | 1.47 | 0 | 0 | 1.475 | 0 | 0.13275 | 0 | 132.75 | 0 |
All samples look good for DNA and RNA concentrations except for 491. It had a low DNA concetration and no RNA was detected, even though I used the High Sensitivity kits for both assays.
Gel
I ran a 1.5% gel at 100 volts for 1 hour. Samples are in the same order as the Qubit with an empty well in between the ladder and the samples.
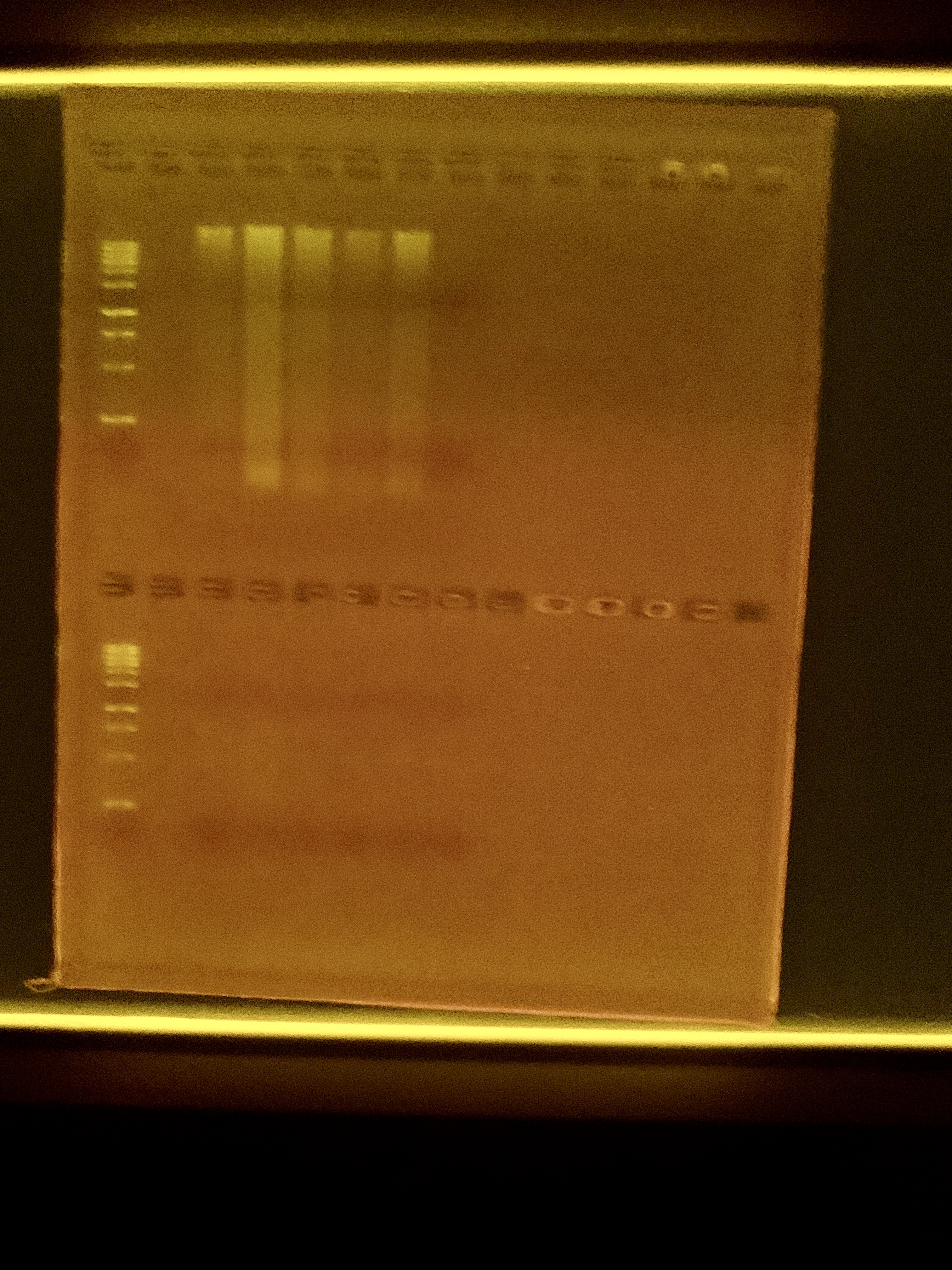
There is DNA there, some degradation. No DNA for 491. No RNA showed up, which was weird. Could be degradation or gel loading issues.