Lipid assay
20230915 Lipid assay test
This protocol was modified from Bove & Baumann 2021 and L. Fuess lipid protocol. We are testing the lipid assay with coral eggs, embryos, and larvae.
The samples used in this assay are test physiology samples from my developmental time series project with M. capitata in Hawaii 2023. For experimental details, see my Hawaii notebook posts here.
Lauren and I followed this protocol with a few modifications. I am going to review the steps in this post and add more details/pictures.
Protocol
Remember to do this protocol in fume hood whenever possible! Reagents are very toxic!
Make reagents according to the linked post above
Lipid assay
- Prior to the assay, homogenize the eggs/embryos/larvae in 700 uL of PBS.
- Separate out 300 uL of holobiont fraction into a new tube.
- Centrifuge for 5 minutes at 3000 rpm to separate host and symbiont fraction.
- Move the supernatent (host) into a new tube.
- Resuspend pellet (symbiont) with 300-400 uL of 1x PBS.
- In past attempts, we found that 150 uL of sample extract was too concentrated. We decided to do a dilution to see what volume of sample would be best to get a good signal. We did the following:
- 10 uL of sample + 140 uL DI water
- 25 uL of sample + 125 uL DI water
- 50 uL of sample + 100 uL DI water
- Desiccate samples overnight in a SpeedVac.
This is what the SpeedVac settings should be set at:
- Time - run infinitely or until it’s turned off
- Brake - OFF or no spinning
- Temp - room temp
- Mode vent - D-AQ or desiccate aqueous solution
This is what the samples looked like after dessication:
- Add 500 uL of a 2:1 chloroform:methanol mixture and 100 uL of 0.05 M NaCl to each desiccated sample.
- 333.33 uL of chloroform and 166.67 uL of methanol
- Vortex samples for 15 seconds to dissolve lipids.
- Place samples on a shaker for 1 hour (set shaker to 500 rpm), vortexting the samples for 10 seconds every 15 minutes.
- While the samples are shaking, make the standards:
- Label 7 1.5 mL tubes 1-7.
- Add 300 μL of CHCl3 to standard tubes 2 through 7
- Add 600 μL of 1.5 mg/ml corn oil stock to standard tube 1
- Transfer 300 μL of tube 1 and place in tube 2. Pipette mix
- Pull 300 μL from tube 2 and place in tube 3. Pipette mix
- Repeat this process for tubes 3 through 6. Do not add corn oil to tube 7! This is the blank
- Discard 300 μL from tube 6 so total volume equals 300 μL (optional)
| Standard | Concentration (mg/mL) |
|---|---|
| 1 | 1.5 |
| 2 | 0.75 |
| 3 | 0.375 |
| 4 | 0.188 |
| 5 | 0.094 |
| 6 | 0.047 |
| 7 (Blank) | 0 |
- Centrifuge samples at 3000 rpm for 5 minutes.
- Move 100 uL of the standard or bottom layer of the centrifuged sample to a 96-well PCR plate in duplicate.
This is the plate map:

- Add 50 uL of methanol to each well.
- Put plate in hot bead bath (70°C) for 15 minutes.
- Bead bath is essentially the same as a water batch except it’s aluminum beads instead of water.
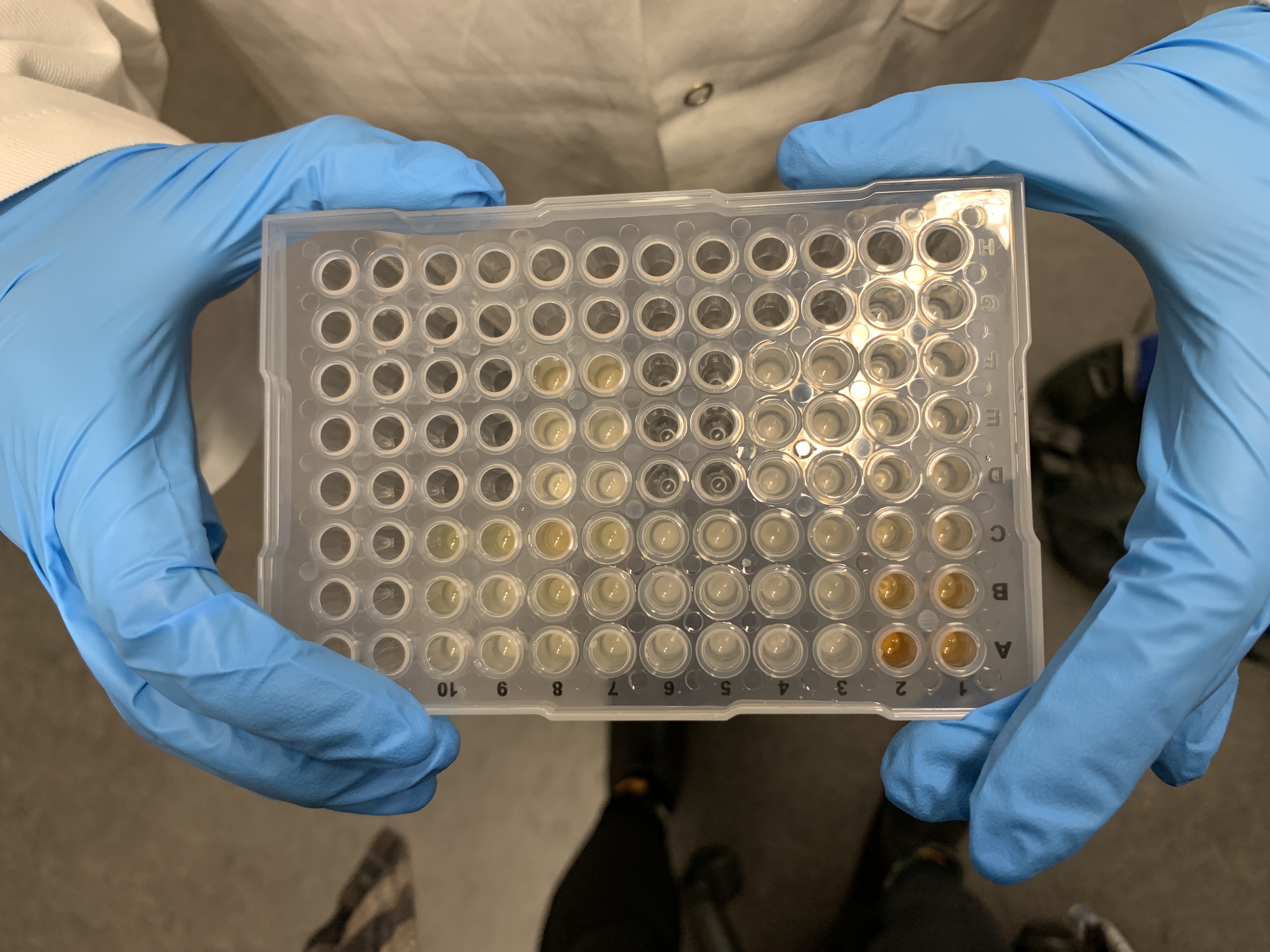
- Following solvent evaporation, add 100 uL of 18 M sulfuric acid to each well. Add the sulfuric acid slowly, as it tends to bubble and splash as it comes into contact with the sample. If the sample splashes into another well and contiminates another sample, toss that plate and re-start.
This is what the PCR plate looked like post-acid addition:
- Put the PCR plate in a thermocycler at 90°C for 20 minutes, followed by 4°C for 20 minutes.
- Transfer 75 uL from the PCR well plate into a new 96 well plate (not a PCR plate).
- Run an initial absorbance reading on the plate reader at 540 nm. This is a baseline measurement that will be used for correcting the final plate absorbance.
This is what the lipid plate looked like before the initial reading:
These are the values from the initial reading:
- Remove the plate from the plate reader and add 34.5 uLof 0.2 mg/mL vanillin in 17% phosphoric acid to each well.
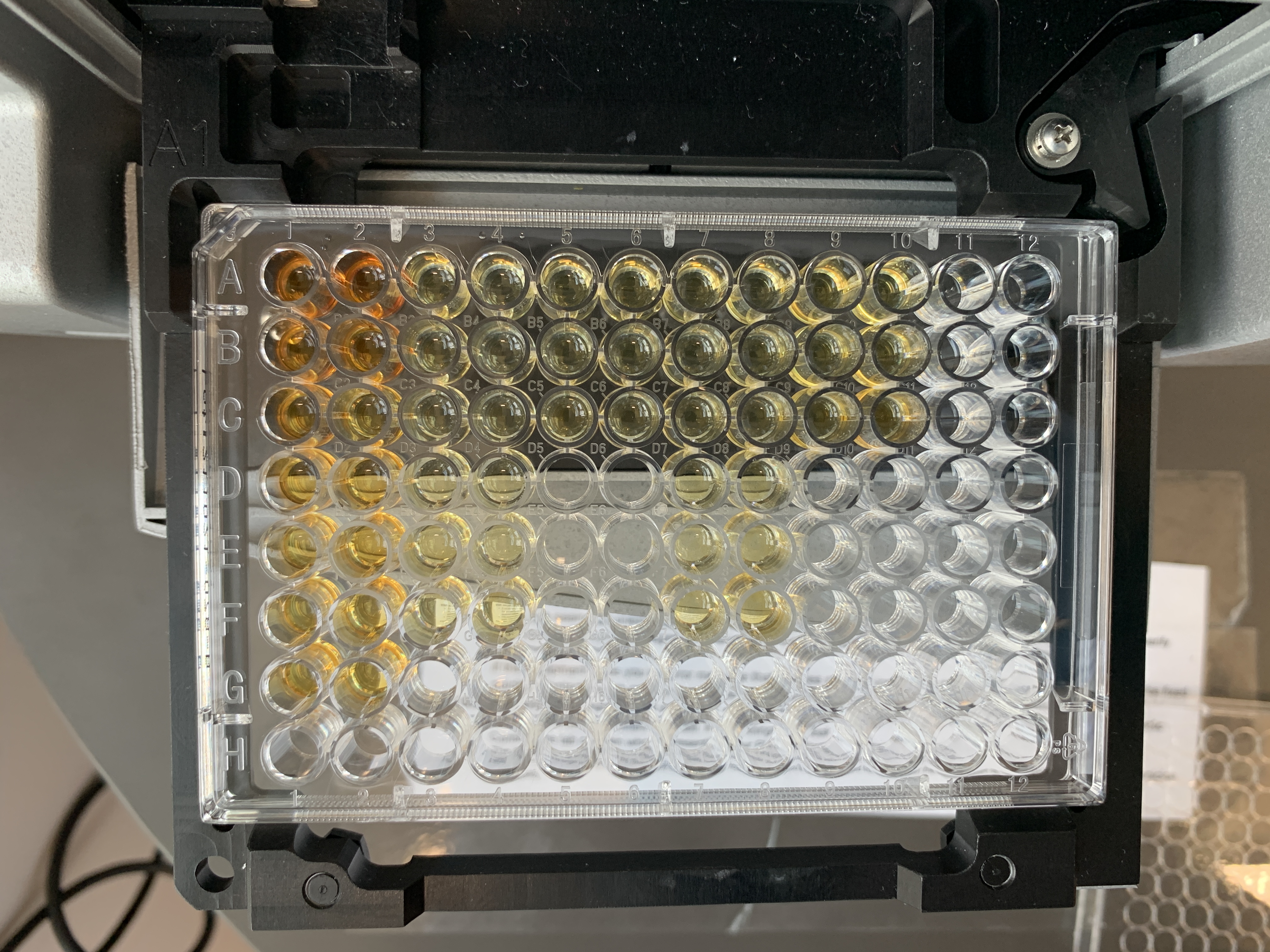
- Incubate the plate in the dark at room temperature for 5 minutes.
- Read the plate again at 540 nm with the lid off.
This is what the lipid plate looked like after the incubation and before the final reading:
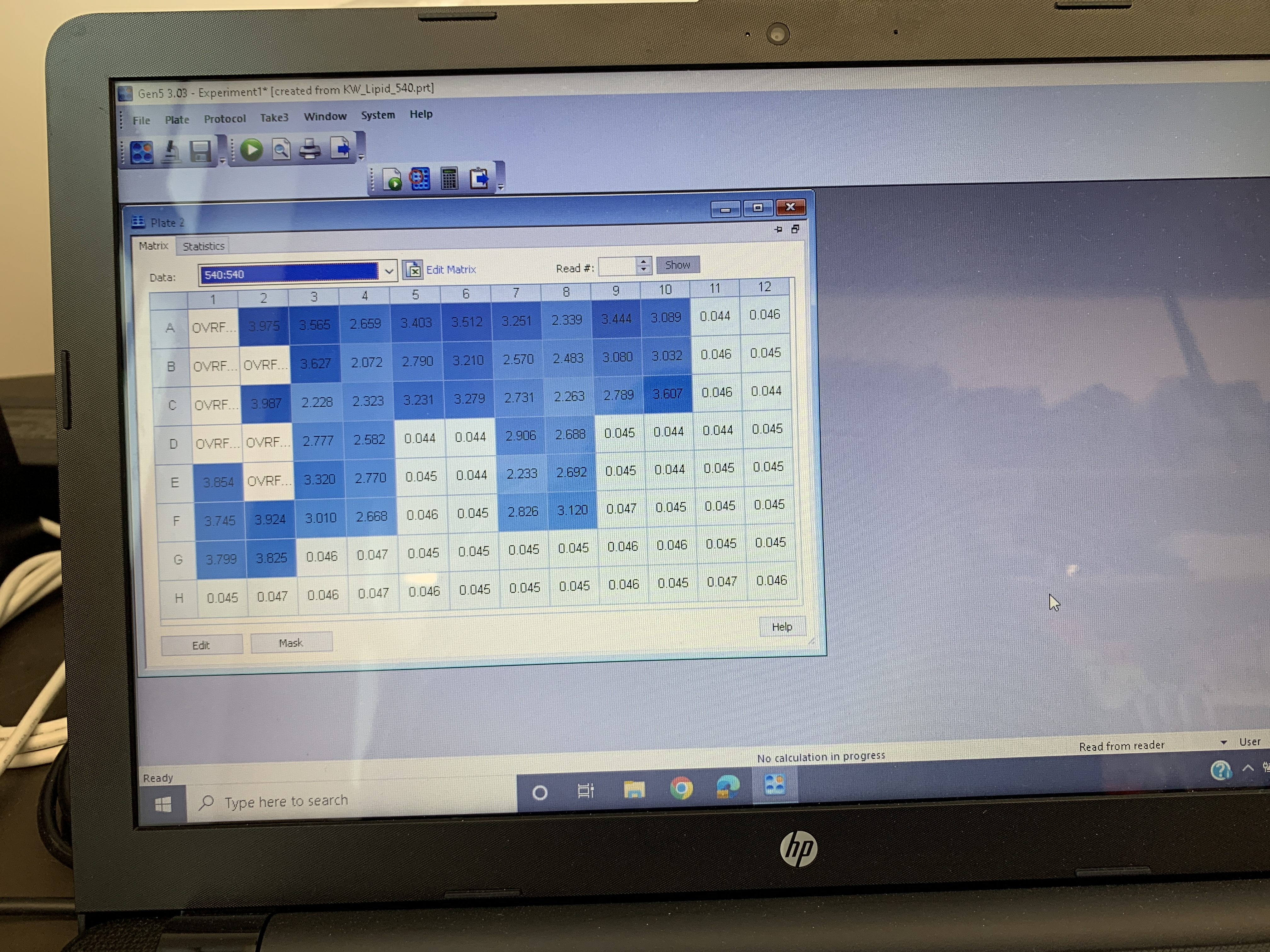
These are the values from the final reading:
Similarly to previous attempts, the values were really high, but only for the standards.
We used a 2:1 chloroform:methanol mixture to extract lipids following Cheng et al. 2011 and Bove & Baumann. This is what Cheng et al. 2011 plate looked like following the vanillin/phosphoric acid addition:
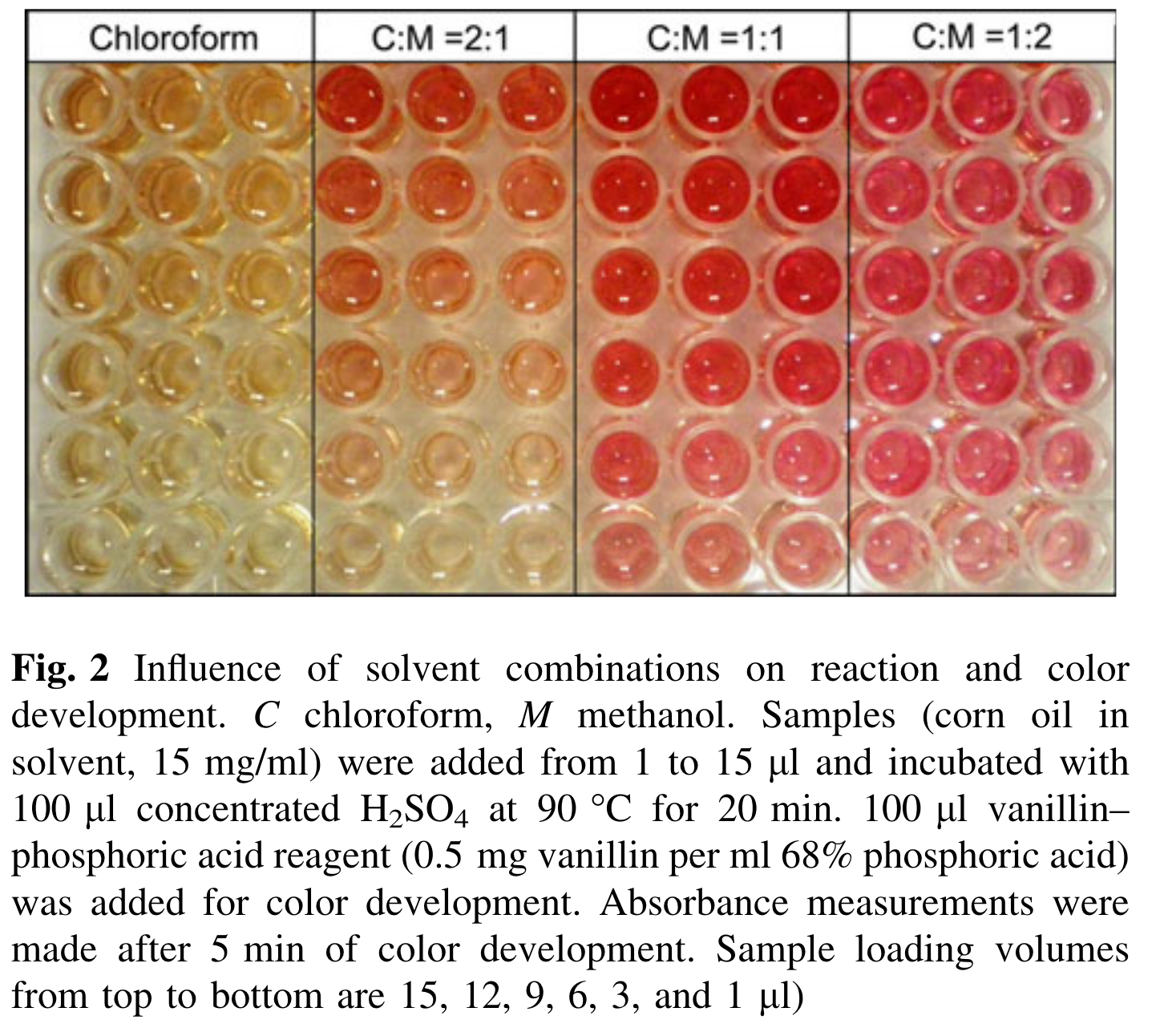
Ours looked darker, more similar to the C:M 1:1 from their paper. They also used a higher vanillin and phosphoric acid ratio (they used 0.5 mg vanillin per mL in 68% phosphoric acid, while we used 0.2 mg vanillin per mL in 17% phosphoric acid).
Next steps
- Check expiration dates on all reagents to make sure nothing is expired
- Discuss at lab meeting
- Maybe remake the vanillin in phosphoric acid?