ITS2
ITS2 for Mcap developmental timeseries Hawaii 2023 and POC spawning 2023 sample
This post details information on ITS2 amplification for the my ambient Mcap developmental timeseries samples from 2023 and for the POC spawning experimental samples from 2023. Mcap 2023 github repo is here and POC github is here.
These samples had pretty low DNA concentrations (see DNA QC post here for sample concentration info). In this test, I attempted to dilute 6 samples to the lowest concentration (0.104 ng/uL) and run the PCR amplification.
I followed Ariana’s ITS2 protocol with some modifications for testing some samples with low concentrations.
Samples
There are 44 samples total to process for ITS2 but today, I only used 6 as a test. Samples used today were:
- Positive control (R55 from AH Mcap 2023 experiment)
- Negative control (ultrapure h2o)
- M9
- M51
- M83
- 6
- 109
- 112
Samples were thawed on ice for dilutions.
Equipment and materials
- 2X Phusion Mastermix
- Forward primer (ITSintfor2) at 100uM: TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG GAA TTG CAG AAC TCC GTG
- Reverse primer (ITS2_Reverse) at 100uM: GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGG GAT CCA TAT GCT TAA GTT CAG CGG GT
- Loading Dye NEB 6X Purple Loading Dye NEB Cat # B7024S
- Gel Stain Biotium GelGreen Nucleic Acid Gel Stain, 10,000X in Water Fisher Cat NC9728313
- DNA Ladder 1kb Gel ladder and 100bp gel ladder
- Agarose and 1XTAE buffer for gel electrophoresis
- Ultrapure water
- PCR strip tubes and 1.5 mL tubes
Protocol
Before starting the protocol, prepare the following:
-
Sample list. If you are performing preparations in multiple batches/PCRs, randomize the sample order for processing to remove batch effects confounding with treatment groups.
-
Prepare list of appropriate controls. Controls will include a negative control (input = nuclease-free water) and a positive control (gDNA from a previously successfully amplified and sequenced sample). These controls should be included in each batch. If you perform preparations over multiple days, include a positive and negative control each day or with each PCR.
- Label 3 PCR strip tubes per sample (including negative and positive controls) with the sample ID
- Since I was only running a test today, I just had 1 replicate per sample instead of 3
- 96 well PCR plates can also be used for lots of samples
- Labeled 1 PCR strip tube per sample for pooled PCR product
Sample Dilution
First, dilute gDNA from each of your samples to a consistent concentration. ITS2 amplifies readily, so input DNA into the first PCR can be at a low concentration. We dilute samples so that there is reduced bias towards high DNA concentration samples during PCR and subsequent sequencing. My lowest DNA concentration was 0.104 ng/uL so I will attempt to dilute all of my samples to that concentration.
- Calculate the volume required to obtain ~0.1 ng/uL gDNA in a 11.5 uL total volume.
- This step is specific to my samples. Usually, DNA is diluted in 10 uL
- Thaw gDNA on ice
- Aliquot the required volume of water for each sample into the labeled gDNA strip tubes.
- For each sample, aliquot the required volume of gDNA into the labeled gDNA strip tubes
- After adding gDNA and water to each tube, vortex for ~5 sec and then spin down.
- Samples can be stored at -20°C until proceeding to the next step.
Here are the samples that I did today:
| TubeID | hpf | Treatment | Project | Qubit HS (ng/uL) | DNA for dilution (uL) | Water (uL) | Total volume | Final concentration (ng/uL) |
|---|---|---|---|---|---|---|---|---|
| R55 (POS) | Larvae | Cladocopium; Ambient | Mcap 2023 AH | 14.3 | 0.50 | 11 | 11.50 | 0.6217391304 |
| Ultrapure H2O (NEG) | NA | NA | NA | NA | 0.00 | 11.5 | 11.50 | 0 |
| M9 | 1 hpf | Ambient | Developmental Timeseries 2023 | 0.104 | 11.50 | 0 | 11.50 | 0.104 |
| M51 | 22 hpf | Ambient | Developmental Timeseries 2023 | 3.53 | 0.50 | 11 | 11.50 | 0.1534782609 |
| M83 | 72 hpf | Ambient | Developmental Timeseries 2023 | 19.45 | 0.50 | 11 | 11.50 | 0.8456521739 |
| 6 | Embryos | Ambient | POC | 0.131 | 11.50 | 0 | 11.50 | 0.131 |
| 109 | Larvae | High | POC | 2.77 | 0.50 | 11 | 11.50 | 0.1204347826 |
| 112 | Larvae | Ambient | POC | 1.71 | 0.61 | 10.89 | 11.50 | 0.09070434783 |
The smallest pipette we have is 0.5, so I had to round up for some samples, which made the concentrations a little all over the place (but all still <1 ng/uL). I could do a serial dilution in the future.
Make master mix
Master mix will be prepared to allow for triplicate reactions for each sample. Reactions are performed in triplicate to 1) increase total PCR product for each sample, 2) provide individual replicates in the case that some reactions do not amplify or do not amplify well and 3) increase diversity of sequences amplified by chance. For the test today, I did NOT run my samples in triplicate.
- Prepare master mix components. Thaw the Phusion Master Mix on ice. Thaw the primers on ice. If required, hydrate lyophilized primers if not done so already. Thaw nuclease-free water on ice. Thaw diluted gDNA samples on ice.
- Calculate the volume of each regent needed by first calculating the total number of reactions requires. The total reactions will be the number of samples (including positive and negative control) x 3. Include an extra 10% volume for pipetting error.
- Once again, only doing 1 replicate today so my total number of reactions + error is 9.
- Label a 1.5 mL tube for master mix
- Label two 1.5 mL tubes for dilution of primers (if required). This protocol calls for 10uM primers. Primers are often received or hydrated at 100uM. To dilute, add 1 part primer to 9 parts nuclease free water. For example, to make 10 total uL of primer stock, add 1 uL of the respective primer to 9 uL of H20. See the table below for the calculation for total primer stock needed to make the master mix. Do this for both the F and R primer separately. Vortex and spin down after diluting.
- Add in volume of Ultra pure/ nuclease free H20 required to the master mix tube. I did NOT do this today, as I added 11.5 uL of diluted DNA to the tube. The protocol typically requires 10.5 uL of ultrapure water per reaction and 1 ng of sample DNA.
- I decided to increase it today because my concentrations were so low.
- Add the necessary volume of Phusion master mix to the 1.5 mL tube
- Add the necessary 10 uM F and R primers to the 1.5 mL tube
Master mix calculations for today:
| Per rxn (uL) | Rxns | Total volume (uL) | |
|---|---|---|---|
| 2x Phusion Master Mix | 12.5 | 9 | 112.5 |
| F Primer (10um) | 0.5 | 9 | 4.5 |
| R Primer (10um) | 0.5 | 9 | 4.5 |
| Ultrapure H2O | 0 | 9 | 0 |
| DNA | 11.5 | ||
| TOTAL VOLUME PER RXN | 25 |
- Add 13.5 uL of master mix to the 11.5 uL of DNA sample
- Vortex and spin down.
- Keep tubes on ice. Samples are now ready for PCR
PCR
Conduct PCR to amplify the ITS2 region. Load all reaction tubes (including negative and positive controls) to the thermocycler. Run the following program (pre-programmed “ITS2” PCR protocol under PUTNAM):
- 1 cycle at 95°C for 3 min
- 35 cycles at 95°C for 30 sec, 52°C for 30 sec, and 72°C for 30 sec
- 1 cycle at 72°C for 2 min
- Hold at 4°C
PCR will run for approx. 1.5 hours. Samples will stay at 4°C on the thermocycler until running a QC gel or store samples at 4°C in fridge overnight.
This is a good stopping point if needed.
Gel QC for PCR product
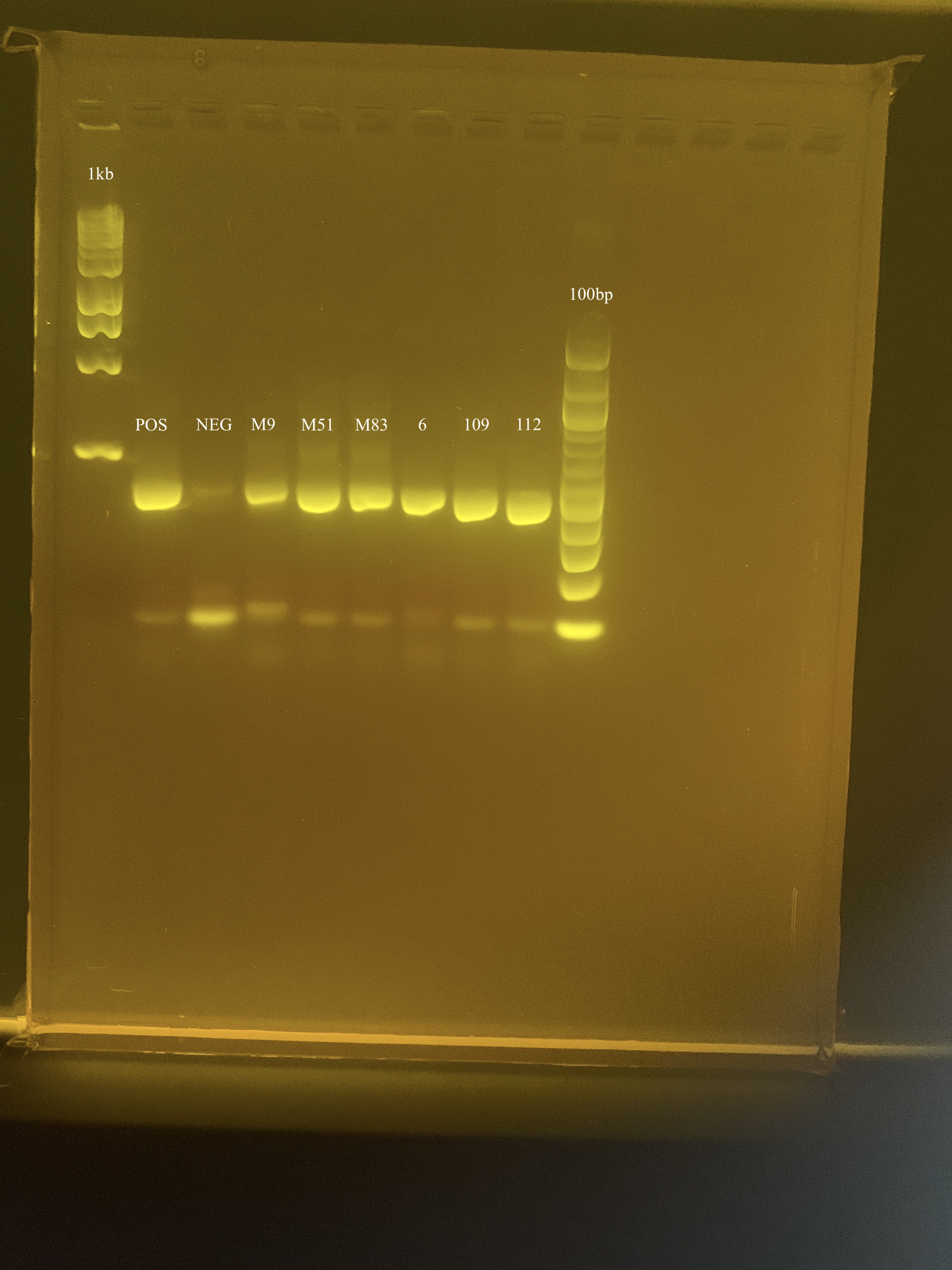
Run a 2% gel at 80-100V and 100 amps for 1.5 hours. Use the 1 kb DNA ladder and 100 bp ladder to each row.
Everything amplified with minimal primer dimer! There is some contamination in the NEG sample. When processing the rest of the samples, I will make new primer dilutions and use the new master mix. Now that I know the amplification can work with low DNA concentration, I will process the samples in triplicate.